We have two funded postdoctoral positions available. Topics encompass:
-Next gen light-sheet fluorescence microscopy instrumentation
-Structured illumination microscopy and other approaches to extend the resolution limit.
-Nonlinear microscopy combined with adaptive optics / phase conjugation
27.01.2026 22:29 — 👍 51 🔁 25 💬 3 📌 2
Fullfabric :: AITHYRA-CeMM PhD
I'm recruiting 1-2 grad students through the AITHYRA-CeMM PhD program! Applications are due January 30th. This is a fully-funded PhD program, combining AI and biology to advance biological discovery. Please forward to anyone who may be interested! You can apply here: apply.cemm.at
08.01.2026 10:48 — 👍 6 🔁 10 💬 1 📌 0

Whiteboard with a drawing of several bears. One bear is thinking, “where am I? I feel so lost down here.”
Some new art popped up in our microscope basement. Big fan
16.01.2026 12:43 — 👍 1 🔁 0 💬 0 📌 0

Direct labeling of microtubule turnover reveals in-lattice repair and stabilization patterns in developing neurons
The microtubule cytoskeleton is the backbone of neuronal morphogenesis, driving the development of the dendrites and axon, and supporting trafficking to distant compartments. How neuronal microtubules...
🚨The Neurocyto lab is branching out in our latest preprint! We used tubulin microinjection to directly visualize microtubule turnover in developing hippocampal neurons, demonstrating the presence of in-lattice repair and a selective stabilization in the nascent axon. Check below, or read on 🧵 1/9
12.01.2026 19:14 — 👍 102 🔁 31 💬 4 📌 2

Dear Sir Paul,
Re: Royal Society Code of Conduct
I am sure that many scientists have written to you about the specific question of Elon Musk’s Fellowship and whether, under the Royal Society’s Code of Conduct, his retaining that Fellowship is appropriate. I will not rehash these issues. Instead, as a female scientist with extensive experience of activities aiming to increase equality, diversity and inclusion in the engineering and physical sciences sector, I am writing to you (in a personal capacity) to ask you to reconsider the statements you have recently made in this context to the UK press about the Royal Society’s Code of Conduct and how it is applied.
A 2018 report from the joint National Academies of the United States of America, concluded that “sexual harassment is common in academic science, engineering, and medicine” and that “greater than 50 percent of women faculty and staff and 20–50 percent of women students encounter or experience sexually harassing conduct in academia”. This report described codes of conduct that make clear that sexual harassment is unethical and will not be tolerated as a “powerful incentive for change”. The authors also noted that sexual harassment can have significant and damaging effects on the integrity of research. In my own praxis, I have found that clear and consistently-implemented codes of conduct that address these issues make female scientists and engineers safer, and allow them to focus more effectively on their research. For codes of conduct to have such a positive effect, it is vital that sanctions for actions which transgress the code are meaningful and substantial.

I was hence aghast to realise that in an interview with the Financial Times published on 9/1/26, you appear to have suggested that the Royal Society “should only expel fellows if their science proved “faulty or fraudulent or highly defective””. Moreover, in a further interview with the Guardian on 11/1/26 you suggested that the code “may need to be looked at again”, with the implication that your aim would be to remove the option of sanctions on Fellows for reasons not strictly related to faults or defects in their research.
I suggest that changing the Royal Society’s code of conduct so that the likelihood of serious sanctions for sexual harassment is reduced, would directly endanger women who interact with the Royal Society at events or otherwise, and would provide a licence to harass to the already powerful people on whom the Society bestows fellowship. The implications of your words - that under your leadership the only infringements of the code which are likely to receive the sanction of the Fellowship being removed are those related to research misconduct - already risk empowering harassers. You stated, in the Financial Times interview, that “there’s many bad people around, but they have made scientific advances”. Given this awareness of the possibility of bad actors in our scientific community, it is wholly irresponsible to suggest that the Royal Society would not act to sanction these people if they harass more vulnerable scientists.
I am hence writing to request that you retract any suggestion that the Society’s Code of Conduct should be changed so that the only reason a Fellow might be sanctioned by the removal of their Fellowship is “faulty or fraudulent or highly defective” research. This action is necessary to safeguard female scientists, a requirement placed on the Society by safeguarding legislation and UK statutory guidance.
Yours sincerely,
Professor Rachel A. Oliver.
Following coverage over the weekend of Sir Paul Nurse's comments that suggested that the only reason that a Fellow should be expelled from @royalsociety.org is scientific misconduct, I have written to him to explain the risks such an attitude poses of increasing sexual harassment in STEM.
12.01.2026 08:59 — 👍 813 🔁 298 💬 25 📌 29
Manim (www.manim.community) is such a neat tool for generating videos that describe microscopy theory. It’s possible to animate a fluorophore behaving as a Lorentzian with a single line of code—amazing! Code (I’m so sorry about the variable names) here: github.com/zacsimile/ra...
12.01.2026 10:04 — 👍 17 🔁 3 💬 0 📌 0
Coffee stains: not just for furniture
10.01.2026 13:13 — 👍 3 🔁 0 💬 0 📌 0
It’s an interesting argument: “If you want to remove Musk from the Royal Society, you better be prepared to remove other scientists who have exhibited bad behavior.” I think that sounds fine.
10.01.2026 12:30 — 👍 8 🔁 0 💬 1 📌 0
Now out in @natcomms.nature.com: www.nature.com/articles/s41...! The latest GitHub commits/Colab notebooks enable simulation with 3D sequences.
09.01.2026 12:59 — 👍 7 🔁 3 💬 0 📌 0

4Pi-SIMFLUX: 4Pi single-molecule localization microscopy with structured illumination - Nature Methods
4Pi-SIMFLUX is a single-molecule localization microscopy approach that achieves a near-isotropic resolution below 10 nm in whole mammalian cells.
We are happy to share our latest work, 4Pi-SIMFLUX, which combines structured illumination with interferometric detection to achieve near-isotropic 3D localization precision of 2–3 nm and resolve sub-10 nm structural features across whole mammalian cells.
www.nature.com/articles/s41...
rdcu.be/eQVxt
21.11.2025 03:00 — 👍 29 🔁 8 💬 0 📌 0

We present multi-immersion Oblique Plane microscope (miOPM), a light-sheet platform that can be adapted to a wide range of applications, from sensitive live cell imaging to imaging organs and cleared tissues.
www.biorxiv.org/content/10.1...
06.10.2025 17:52 — 👍 123 🔁 44 💬 5 📌 2

Localization of single molecules with structured illumination and structured detection - Light: Science & Applications
Light: Science & Applications - Localization of single molecules with structured illumination and structured detection
Happy to share this article with my views on @elislenders.bsky.social and @vicidominilab.bsky.social work and discussing current developments in single-molecule localization combining structured excitation and detection. So many exciting perspectives in the field!
www.nature.com/articles/s41...
30.09.2025 18:17 — 👍 23 🔁 10 💬 2 📌 0
Highly efficient 12-color multiplexing with speed-optimized DNA-PAINT. We are excited to share our latest paper in @natcomms.nature.com, using left-handed DNA to extend speed-optimized DNA-PAINT to 12 targets in a simple and straightforward way! 🧬👈🚀https://www.nature.com/articles/s41467-025-64228-x
02.10.2025 11:37 — 👍 28 🔁 9 💬 2 📌 2

Sketches of six stages of cytokinetic development of the intercellular bridge are illustrated with exmaple U-ExM fluorescence microscopy images. Septin2-GFP shown in green. Tubulin shown in magenta. A shortened workflow illustrates the image alignment and averaging process to generate average 2D reconstructions of cytokinetic stages.
It is out! 🦚 I recorded hundreds of #ExM 🔬images of cytokinetic bridges and averaged them into 6 stages. How? With help from our fantastic collaborators @zacsimile.bsky.social and @jonasries.bsky.social in Vienna 😇. Check out the full atlas here: doi.org/10.5281/zenodo.17232370
30.09.2025 16:24 — 👍 32 🔁 11 💬 1 📌 1
Now out on bioRxiv. 🥳My research on #cytokinesis, averaging thousands of #ExM images🔬, creating a dynamic atlas of cytokinesis 🦠⏳. Here's an animated sneak peek of what we found. Better resolution on bioRxiv😄 #PSFoftheGIF
28.09.2025 14:15 — 👍 69 🔁 26 💬 2 📌 4
Automated optogenetic control of hundreds of cells in parallel. Each cell is individually steered, collectively acting as a "tissue printer". Preprint & code out! www.biorxiv.org/content/10.1...
21.08.2025 20:16 — 👍 110 🔁 38 💬 6 📌 2

🚨 2 × PhD positions @EPFL! 🚨
Help us push the boundaries of fluorescence microscopy - DNA nanotech, custom optics & spatial omics in Lausanne 🇨🇭. Start Jan 2026. Send CV + motivation + 2 refs → fschueder@ethz.ch
#PhD #Hiring #microscopy #SuperResolution #SpatialOmics #DNAPAINT #FLASHPAINT
31.07.2025 08:56 — 👍 24 🔁 20 💬 1 📌 1

Scientists hide messages in papers to game AI peer review
Some studies containing instructions in white text or small font — visible only to machines — will be withdrawn from preprint servers.
Human scientists anticipate human reviewers will use machines, so they put tiny white text to prompt machines to give positive reviews. I’m not even angry, I’m impressed www.nature.com/articles/d41... Scientists hide messages in papers to game AI peer review
14.07.2025 07:38 — 👍 39 🔁 8 💬 5 📌 2

A picture of an ice maker full of ice cubes
Good news I've fixed the worst thing about Europe
29.06.2025 16:56 — 👍 5 🔁 1 💬 1 📌 0
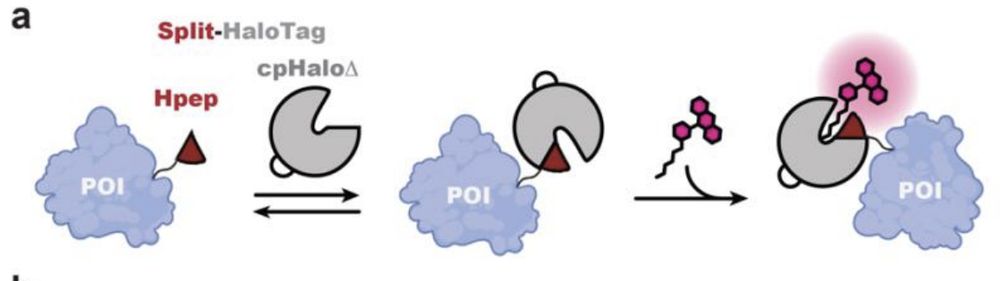
schematic illustration of protein labeling using the spontaneous self-complementing peptide protein split-HaloTag system. POI: protein of interest

Pairwise combination of Hpep11 (TOM20-tagged) with the other three Hpep variants 8, 9 and 10 (H2B-tagged) for FLIM multiplexing. The total fluorescence intensity composite, the two separated species and their corresponding wavelet-filtered phasor plot used for species separation are presented.
![Live-cell confocal imaging of histone H2B type-2E-Hpep11 CRISPR KI cell lines after 2-hours labeling with the specified CA- ligand [100 nM]. Images were taken with optimal image acquisition parameter for each dye. Scale bar: 50 μm.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:uf4m7zlmcjlg2biwn4s2k32y/bafkreibozn5fz6sgeksqrz3vmy6uiydtnvwmb5my7r3xecko2erokryn3a@jpeg)
Live-cell confocal imaging of histone H2B type-2E-Hpep11 CRISPR KI cell lines after 2-hours labeling with the specified CA- ligand [100 nM]. Images were taken with optimal image acquisition parameter for each dye. Scale bar: 50 μm.
![(a) Confocal laser scanning microscopy (CLSM) and STED images of mitochondria in U2OS cells coexpressing cpHaloΔ3 and TOM20-Hpep, either overexpressed or endogenously tagged. Scale bar: 1 μm. Pixel intensities scaled according to reference bar. (b) Representative CLSM, STED images of the CRISPR/Cas9 KI cells expressing TOM20 tagged with intact HaloTag (upper) or Hpep11 (bottom). (c) Intensity profiles along mitochondrial tubules (red and blue lines in b). Scale bar: 2 μm. (d) Representative CLSM and STED images of endogenous Hpep11-tagged clathrin with cpHaloΔ3 coexpression. Scale bar: 10 μm (overview) and 2 μm (magnification). (e) Representative CLSM, STED
images of endogenously tagged tubulin beta 4B with Hpep11. Scale bar: 10 μm (overview) and 2 μm (magnification). (f) Intensity profiles along tubulin filaments (red and blue lines in e) Means ± s.d. of the filament diameters were calculated as full width at half maximum (FWHM) from n=20 microtubule filaments, ≥ 2 images. A slight increase in cytosolic signal was noted in cells tagged with split-HaloTagat TUBB4B, compared to cells tagged with the full-length HaloTag, which may result from the presence of unbound but labeled cpHaloΔ3. All images were acquired after labeling with CA-SiR [100 nM] for one hour.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:uf4m7zlmcjlg2biwn4s2k32y/bafkreielty6fex37xy4caechbw5ebo3eypyiyvxxhkigeazd5gmy6tvlay@jpeg)
(a) Confocal laser scanning microscopy (CLSM) and STED images of mitochondria in U2OS cells coexpressing cpHaloΔ3 and TOM20-Hpep, either overexpressed or endogenously tagged. Scale bar: 1 μm. Pixel intensities scaled according to reference bar. (b) Representative CLSM, STED images of the CRISPR/Cas9 KI cells expressing TOM20 tagged with intact HaloTag (upper) or Hpep11 (bottom). (c) Intensity profiles along mitochondrial tubules (red and blue lines in b). Scale bar: 2 μm. (d) Representative CLSM and STED images of endogenous Hpep11-tagged clathrin with cpHaloΔ3 coexpression. Scale bar: 10 μm (overview) and 2 μm (magnification). (e) Representative CLSM, STED
images of endogenously tagged tubulin beta 4B with Hpep11. Scale bar: 10 μm (overview) and 2 μm (magnification). (f) Intensity profiles along tubulin filaments (red and blue lines in e) Means ± s.d. of the filament diameters were calculated as full width at half maximum (FWHM) from n=20 microtubule filaments, ≥ 2 images. A slight increase in cytosolic signal was noted in cells tagged with split-HaloTagat TUBB4B, compared to cells tagged with the full-length HaloTag, which may result from the presence of unbound but labeled cpHaloΔ3. All images were acquired after labeling with CA-SiR [100 nM] for one hour.
New preprint by @kjohnsson.bsky.social lab!
A new split Halotag system with higher affinity of the complements + lower background. The system works with our SiR-CA & CPY-CA halotag ligands and enables STED imaging or FLIM multiplexing.
The short 14 aa tag Hpep enables easy cloning free CRISPR-KI.
17.06.2025 11:09 — 👍 28 🔁 4 💬 2 📌 0


Tiny yellow bird waiting at the metro stop this morning
25.04.2025 06:05 — 👍 5 🔁 0 💬 0 📌 0
Your yearly reminder to acknowledge the core facilities you use and their staff scientists in your papers. These scientists are a crucial part of the scientific ecosystem and to continue to exist they need tangible credit for their work. Plus their associated expertise adds credibility to your work.
10.04.2025 13:38 — 👍 488 🔁 165 💬 7 📌 16
Thanks! Yep, we do 3D excitation (z scan with a top hat) and 3D modeling of fluorophore positions. See, e.g., example2.
10.04.2025 05:50 — 👍 1 🔁 0 💬 0 📌 0
Scientist 👩🔬 & EPFL Prof 🇨🇭 | DeepLabCut.org , 🦓 cebra.ai | neuroscience & ML 🧠 mackenziemathislab.org | ✨CSO at Kinematik.ai | occasionally 🐦⬛birds/🌱outdoors/🍣food/👠fashion
Editor in Chief, MeidasNews. Former Fed & State Prosecutor, Marine, Attorney. Switched parties from R to D in 2020. MeidasPlus.com/subscribe
Tour, On Cinema at the Cinema, Office Hours, find it all and more at linktr.ee/tim.heidecker
Chicago's neighborhood news source.
Black-owned publication from Chicago to the 🌍
Reshaping the narrative in pursuit of truth and liberation.
💻: thetriibe.com
📩: https://thetriibe.com/newsletters/
Support us: https://buy.stripe.com/00w14n5ZRbhkbXaaeA6Zy03
Associate Professor, Jiangsu Academy of Agricultural Sciences. CAZymes, filamentous fungi, oomycetes, microbe-microbe & plant-microbe interactions, genomics.
Principal Investigator at @aithyra.bsky.social | Structural bioinformatics, virology, and innate immunity | jasonnomburg.com
Der Wissenschaftsfonds FWF ist Österreichs zentrale Einrichtung zur Förderung der Grundlagenforschung.
www.fwf.ac.at
scilog – das Wissenschaftsmagazin des FWF:
scilog.fwf.ac.at
For students, postdocs and early career professionals, we aim to focus on career development of RMS members through workshops, networking and much more.
Hamamatsu Photonics is a leading manufacturer of devices for visible, infrared, ultraviolet, and x-rays. We are dedicated to enhancing life through light-based technologies.
https://www.hamamatsu.com/
News for international scientists, students and media from Graz University of Technology. #science #passion #technology
Imprint: https://www.tugraz.at/ueber-diese-seite/impressum/
https://nbic.pku.edu.cn/en/Faculty/Researchers/e954671a8e2249e8a1339ab4f42e70ea.htm
Single Molecule Localization and Super resolution microscopy enthusiast @labtinnefeld.bsky.social , LMU Munich
Researcher at the University of Göttingen, Germany. Microscopist, Method developer: super-resolution imaging, SMLM, DNA-PAINT, FLIM, Multiplexing, Microfluidics.
Marie Skłodowska-Curie (MSCA) Postdoctoral Fellow 🇪🇺 | Cell Biologist 🧬 & Super-Resolution Microscopy Specialist 🔬 in the Ries Lab at the University of Vienna working on deciphering apoptosis on the nanoscale.
Formerly in the Jakobs lab at MPI Göttingen.
Independent, reader supported investigative reporting from Chicago on policing and state violence. Occasionally: hate movements and conspiracism.
Support us: unraveledpress.com/support-unraveled/
📧 info@unraveledpress.com
📱 Signal: unraveled.66
Group leader SNSF Ambizione at EPFL, Lausanne. Formerly, Postdoc Roux lab, Geneva; PhD Piel lab, Paris. #cytoplasts #actomyosin #cellbio #microscopy #membranes #biophysics #extracellular_vesicles #sciart
Lab website: http://celldynamicslab.com






















![Live-cell confocal imaging of histone H2B type-2E-Hpep11 CRISPR KI cell lines after 2-hours labeling with the specified CA- ligand [100 nM]. Images were taken with optimal image acquisition parameter for each dye. Scale bar: 50 μm.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:uf4m7zlmcjlg2biwn4s2k32y/bafkreibozn5fz6sgeksqrz3vmy6uiydtnvwmb5my7r3xecko2erokryn3a@jpeg)
![(a) Confocal laser scanning microscopy (CLSM) and STED images of mitochondria in U2OS cells coexpressing cpHaloΔ3 and TOM20-Hpep, either overexpressed or endogenously tagged. Scale bar: 1 μm. Pixel intensities scaled according to reference bar. (b) Representative CLSM, STED images of the CRISPR/Cas9 KI cells expressing TOM20 tagged with intact HaloTag (upper) or Hpep11 (bottom). (c) Intensity profiles along mitochondrial tubules (red and blue lines in b). Scale bar: 2 μm. (d) Representative CLSM and STED images of endogenous Hpep11-tagged clathrin with cpHaloΔ3 coexpression. Scale bar: 10 μm (overview) and 2 μm (magnification). (e) Representative CLSM, STED
images of endogenously tagged tubulin beta 4B with Hpep11. Scale bar: 10 μm (overview) and 2 μm (magnification). (f) Intensity profiles along tubulin filaments (red and blue lines in e) Means ± s.d. of the filament diameters were calculated as full width at half maximum (FWHM) from n=20 microtubule filaments, ≥ 2 images. A slight increase in cytosolic signal was noted in cells tagged with split-HaloTagat TUBB4B, compared to cells tagged with the full-length HaloTag, which may result from the presence of unbound but labeled cpHaloΔ3. All images were acquired after labeling with CA-SiR [100 nM] for one hour.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:uf4m7zlmcjlg2biwn4s2k32y/bafkreielty6fex37xy4caechbw5ebo3eypyiyvxxhkigeazd5gmy6tvlay@jpeg)