The proteasome-substrate-shuttle protein UBQLN2 contains—like other quality control system proteins—a long region devoid of lysine (a lysine desert)
Martin Grønbæk-Thygesen (from @rhp-lab.bsky.social) et al show that introducing K here causes ubiquitylation and degradation
doi.org/10.1101/2025...
07.10.2025 21:19 — 👍 32 🔁 11 💬 2 📌 1

Martin Grønbæk-Thygesen in @rhp-lab.bsky.social measured abundance and toxicity (both independently of function) of missense variants in aspartoacylase, and found that a subset of the low-abundance variants were toxic, induced a stress response which correlated with toxicity
doi.org/10.1038/s414...
19.09.2025 20:57 — 👍 2 🔁 1 💬 0 📌 0

We are back with an exciting seminar on September 8 at 3pm CEST! We’ll hear from Prof. Birthe Kragelund and Maximilian Vieler (@uu.se).
Register to attend:
tinyurl.com/PMCseminar2
Recordings of previous seminars: www.youtube.com/@PMCModularity
15.08.2025 15:35 — 👍 17 🔁 8 💬 0 📌 1
Curiosity underlies a breakthrough in rare disease
We must recognise and protect the pipelines that lead from research to real-world benefit
Both translational and fundamental curiosity-driven research are needed to fuel the incredible progress we're seeing in genomic medicine; an important message in this article and and some lovely quotes from @sarahlwynn.bsky.social
www.ft.com/content/25dd...
29.05.2025 08:44 — 👍 18 🔁 9 💬 0 📌 1
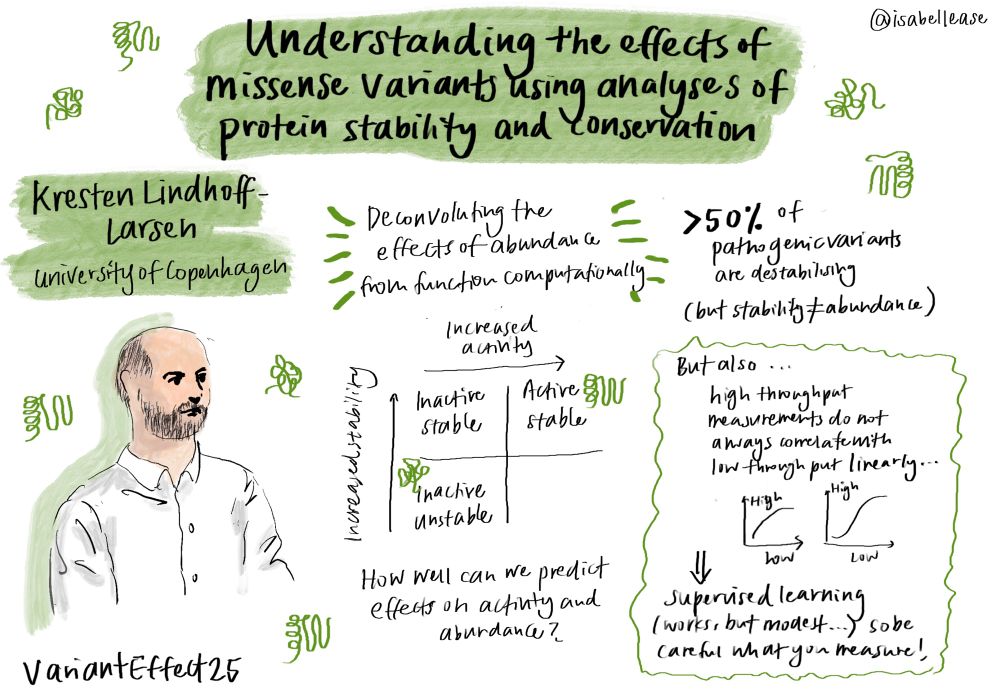
@lindorfflarsen.bsky.social at #VariantEffect25
23.05.2025 08:46 — 👍 22 🔁 5 💬 0 📌 1

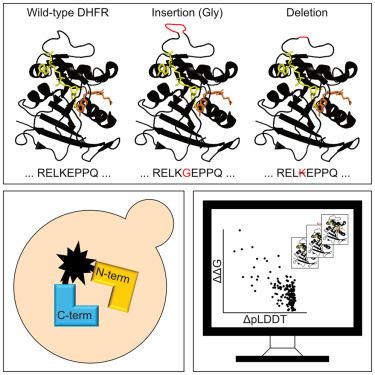
Comprehensive degron mapping in human transcription factors
Gene expression is regulated by the targeted degradation of transcription factors through the ubiquitin-proteasome system. Transcription factors destined for degradation are recognized by E3 ubiquitin...
With the @lindorfflarsen.bsky.social group we present our map of degrons in all human transcription factors, incl. examples of constitutive degrons in exposed regions & buried degrons that are exposed upon mutation. In addition, we show that most TADs overlap with degrons. Work led by Fia Larsen.
18.05.2025 04:59 — 👍 27 🔁 7 💬 0 📌 1

A complete map of human cytosolic degrons and their relevance for disease
Degrons are short protein segments that target proteins for degradation via the ubiquitin-proteasome system and thus ensure timely removal of signaling proteins and clearance of misfolded proteins fro...
In collaboration with the @lindorfflarsen.bsky.social group we release our map of degrons in >5,000 human cytosolic proteins with >99% coverage. A machine learning model trained on the data identifies missense variants forming degrons in exposed & disordered regions. Work led by @vvouts.bsky.social.
15.05.2025 11:56 — 👍 18 🔁 8 💬 0 📌 1
We are looking for a PhD student with interest in protein biochemistry on a really cool project “Cooperation of deubiquitinating enzymes and VCP/p97 in cellular stress responses”. Build your own reactions! fully funded. apply with code 193-25. #ubiquitin
23.04.2025 21:56 — 👍 5 🔁 5 💬 0 📌 0
... wait! There's more great Review content!
☘️ mechanisms of #SAM homeostasis (Ben Tu et al)
☘️ glucose sensing by #glucokinase and how disease variants impact its function @sarahgersing.bsky.social @lindorfflarsen.bsky.social @rhp-lab.bsky.social
06.03.2025 16:43 — 👍 2 🔁 1 💬 1 📌 1
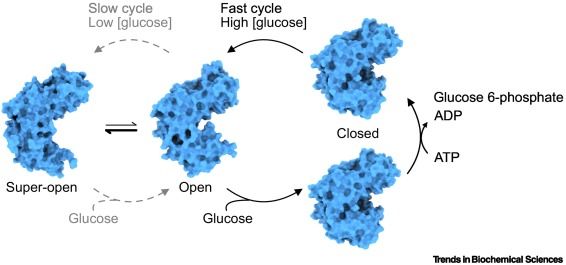
Now online - the Review "#Glucokinase: from allosteric glucose sensing to disease variants" from @sarahgersing.bsky.social, Torben Hansen, @lindorfflarsen.bsky.social, and @rhp-lab.bsky.social.
#GlucoseHomeostasis #GlucoseSensor #GCKMODY #CooperativeKinetics
authors.elsevier.com/a/1kN5M3S6Gf...
03.01.2025 16:40 — 👍 9 🔁 4 💬 1 📌 0
Happy to share our review, 'Glucokinase: from allosteric glucose sensing to disease variants', published in @cp-trendsbiochem.bsky.social, which explores the fascinating enzyme glucokinase. authors.elsevier.com/a/1kN5M3S6Gf...
02.01.2025 19:35 — 👍 20 🔁 2 💬 1 📌 1
The official Bluesky of Naughty Dog, the studio behind Intergalactic, The Last of Us, UNCHARTED, and Jak and Daxter. Games rated RP to M by the ESRB.
Group leader in membrane synthetic biology at ANU, Ngunnawal country. I like cryo-EM, yeast, biosensors and riding bikes. he/him/his.
Uofficiel bot som poster links til dr.dk/nyheder ud fra RSS feed'et, ligesom @bbcnews-world-rss.bsky.social gør for BBC.
Lavet af @janaagaard.com. Hosted på @github.com.
Kildekode: https://github.com/janaagaard75/unofficial-drdk-bluesky-bot.
The best of FT journalism, including breaking news and analysis.
https://www.ft.com
The users this account follows are verified FT staff or contributors.
Else Kröner Fresenius Professor & Chair of Translational Nutritional Medicine @tum.de | PI @helmholtzmunich.bsky.social | Former @lmumuenchen.bsky.social @harvard.edu @uni-hamburg.de | Banner by @albamena.bsky.social
Homepage: https://www.mls.ls.tum.de/tnm
Covering online speech, social media and the information wars at @washingtonpost.com. Shitposting about Bluesky on Bluesky.
Signal: willoremus.24
Professor, Department of Biological Sciences, University of Pittsburgh. RNA polymerase II mechanism, classic films, #classiccountdown once in awhile
Assistant Professor at Baylor College of Medicine and Texas Children's Hospital. Houston, TX
Science: disorder, condensates, repeats, cell stress, neurodegeneration, drug discovery, synbio Non-science: art, fashion, cooking
www.boeynaemslab.org
Associate Professor, Computational biophysicist @UofT
http://rauscher-group.physics.utoronto.ca
Professor at Michigan State University. Trying to understand how the universe works, including people and animals. And plants and microbes. So, pretty much everything. (he/him)
Jumping Genomics!
I run a lab in NYC that studies transposons 🧬in the brain 🧠
Opinions are all mine
mghlab.org
Assistant professor of Biochemistry @CU Boulder. We study Ubiquilins and how dysregulation of the virus-like protein PEG10 contributes to neurodegenerative disease upon UBQLN2 mutation. I'm lucky to lead a great group of scientists in a beautiful place!
Husband, Father and grandfather, Datahound, Dog lover, Fan of Celtic music, Former NIGMS director, Former EiC of Science magazine, Stand Up for Science advisor, Pittsburgh, PA
NIH Dashboard: https://jeremymberg.github.io/jeremyberg.github.io/index.html
Advancing health and well-being in the life sciences through collaborative advocacy. We are the voice of biological/biomedical researchers.🔬
www.faseb.org
https://linktr.ee/FASEBorg
Working for a diverse, global and multidisciplinary scientific community focused on the cell, the basic unit of all life.
Planetary epidemiologist at Yale. Thinking about thinking about thinking about writing a book about viruses. Look, I made a hat: carlsonlab.bio • viralemergence.org • gbcc.study
Historian of technology, & dad joke aficionado who lives w/a tiny, spotlight-stealing rabbit.
I won’t share AI generated content, except to critique it.
If you see me in person I’ll be wearing a mask. If you care abt me, put one on too.
www.marhicks.com
First gen scientist studying protein & RNA ADP-ribosylation as part of a husband-and-wife team!
Current location: Aachen
Past: Zagreb, Oxford, Amsterdam, Maastricht
Mum of 🧒👧
Structural & Systems Biology Lab @ University of Geneva | Exploring protein & proteome assemblies | Structured thoughts, intrinsically disordered views