Structures from AlphaFold3 - while often impressively good - tend to fail representing the dynamic ensembles accurately. And often parts of the structure are not correct.
Adding experimental data, directly in AlphaFold's diffusion step, provides physically realistic protein ensembles. This image shows two cases where AlphaFold3-only structures were largely improved by guiding with experimental data.
📢 New preprint:
Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇
18.10.2025 18:59 — 👍 52 🔁 22 💬 1 📌 0
New IDPSeminars season on deck 🤩
We hope you can join us to learn about some exciting science!
More info in the post below👇
27.08.2025 14:23 — 👍 8 🔁 4 💬 0 📌 0
📢 New preprint: Aromatic Ring Flips Reveal Reshaping of Protein Dynamics in Crystals and Complexes.
We addressed a long-standing question that people debated already in the 1970s - but it still remained open:
Do proteins in crystals move as they do in solution? We used ring flips to find out.
1/n
25.08.2025 04:38 — 👍 45 🔁 17 💬 2 📌 2
We are thrilled to be part of Project Diffuse, building infrastructure for the dynamic future of structural biology.
We are helping to lead the modeling and encoding efforts of this project, including designing infrastructure that allows AI to learn from the full complexity of experimental data.
12.08.2025 17:20 — 👍 9 🔁 3 💬 2 📌 1
PREreview of “Boltz-2: Towards Accurate and Efficient Binding Affinity Prediction”
Authored by Karson Chrispens and 2 other authors
Hi bluesky scientists! Recently, @stephanieaw.bsky.social @fraserlab.com held a workshop at UCSF to discuss methods for modeling heterogeneity in structural biology (conformationalensembles.github.io).
20.06.2025 17:57 — 👍 12 🔁 4 💬 2 📌 0
Very happy that this story found its home at @pnas.org
www.pnas.org/doi/10.1073/...
20.05.2025 18:23 — 👍 35 🔁 4 💬 1 📌 0
I am glad that this profile written by Luc Rinaldi for Toronto Life Magazine is out. It is about what I am doing and where I am going. Thanks to Luc for the time he took to write this piece. #chemsky #compchemsky
23.04.2025 21:25 — 👍 35 🔁 9 💬 1 📌 0
Looking forward to this workshop on Macromolecular Conformational Ensembles in June!
08.04.2025 14:29 — 👍 1 🔁 0 💬 0 📌 0
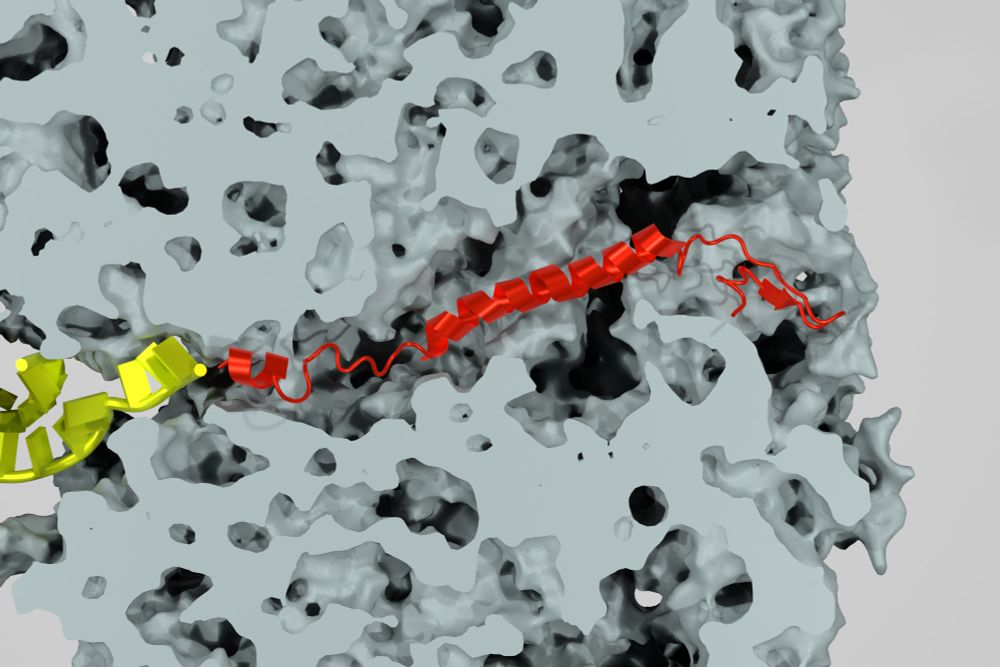
💭 Would you like to contribute to projects in the field of "Theory and Methods for Non-equilibrium #Theory and Atomistic Simulations of Complex# Biomolecules"? Join our department as #PhD student or #Postdoc at @mpi-nat.bsky.social in 🇩🇪 🔗 Here are more details www.mpinat.mpg.de/4965656/10-2... #job
01.04.2025 13:11 — 👍 6 🔁 3 💬 0 📌 1
✨ Check out @mackevinbraza.bsky.social 's new work on full length APOBEC3B, which shows how the NTD modulates active site opening in the CTD
Collab w/ @adaozlemdemir.bsky.social & Aihara, Herzik, & Harris labs
pubs.acs.org/doi/10.1021/...
26.03.2025 15:21 — 👍 57 🔁 11 💬 0 📌 1
This is the moment we've worked toward for a long time: First public disclosure of the @asapdiscovery.bsky.social pan-coronavirus antiviral aiming to help keep humanity safe from future pandemic threats like MERS-CoV and other bat coronaviruses.
25.03.2025 19:47 — 👍 96 🔁 36 💬 4 📌 1
At high concentrations of PEG, we find that mCherry molecules associate to form large aggregates. By carrying out experiments with a bulkier crowder, dextran, we show that the effects of crowder-protein interactions depend on the crowder's chemical features and its bulkiness.
28.02.2025 18:30 — 👍 1 🔁 0 💬 1 📌 0
We find that PEG-wrapping around the side chains of lysine residues is prevalent, occurring in at least 20% of all crystal structures containing PEG, which corresponds to approximately 0.4% of all structures in the PDB.
28.02.2025 18:30 — 👍 2 🔁 0 💬 1 📌 0
We observe a specific, stable PEG-protein interaction in which PEG molecules wrap around the side chains of surface-exposed lysine residues.
28.02.2025 18:30 — 👍 0 🔁 0 💬 1 📌 0
We provide a detailed view of the influence of PEG crowding on the structural equilibrium and the function of a widely used fluorescent protein, mCherry. PEG interacts with the protein non-isotropically, and it is not well-described as a classical excluded volume crowder.
28.02.2025 18:30 — 👍 0 🔁 0 💬 1 📌 0
A recent paper, “PEG-mCherry interactions beyond classical macromolecular crowding,” by Liam Haas-Neill from our lab, Khalil Joron and Eitan Lerner @lernerlab.bsky.social is published in Protein Science.
onlinelibrary.wiley.com/doi/full/10....
28.02.2025 18:30 — 👍 12 🔁 0 💬 1 📌 1
Come to #accelerate25 the premier conference on #aiformaterials #selfdrivinglabs #chemsky Please RT
13.02.2025 21:47 — 👍 17 🔁 13 💬 0 📌 0

Our lab celebrated Ethan Lee's successful PhD defense last week. You can read his recent study on allostery in the main protease of SARS-CoV-2 here: pubs.acs.org/doi/abs/10.1...
Stay tuned for more of his work on IDRs + allostery. Congratulations, Ethan!
13.02.2025 22:44 — 👍 8 🔁 0 💬 0 📌 0
Looking forward to #BPS2025 & presenting our collaborative study with @lernerlab.bsky.social on the effects of PEG crowding on the fluorescent protein mCherry. Drop by our poster (Monday, board B4)!
12.02.2025 22:43 — 👍 12 🔁 0 💬 0 📌 0
It is almost happening - #BPS2025 - and we are happy to attend and present our research.
A thread.
1) I will give a talk on Saturday morning at the Biological Fluorescence subgroup, presenting the work spearheaded by the PhD student Khalil Joron, about
11.02.2025 19:39 — 👍 10 🔁 2 💬 1 📌 0
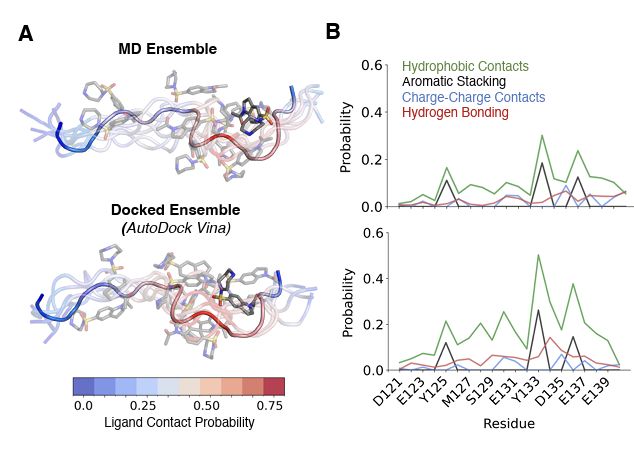
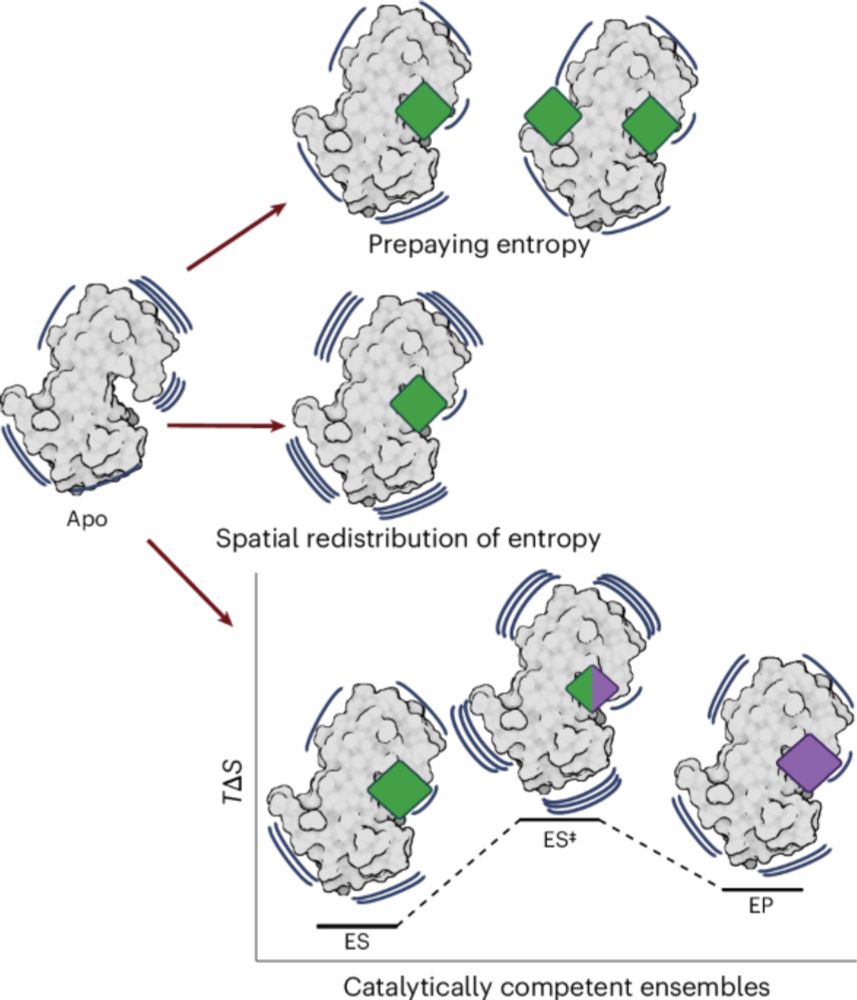
Proud to share:
"Ensemble docking for intrinsically disordered proteins"
from Dartmouth undergrad Anjali Dhar 24' and grad student Tommy Sisk. We present ensemble docking strategies for IDPs that, remarkably, seem to work!
www.biorxiv.org/content/10.1...
Code: github.com/paulrobustel...
31.01.2025 19:58 — 👍 75 🔁 14 💬 2 📌 0
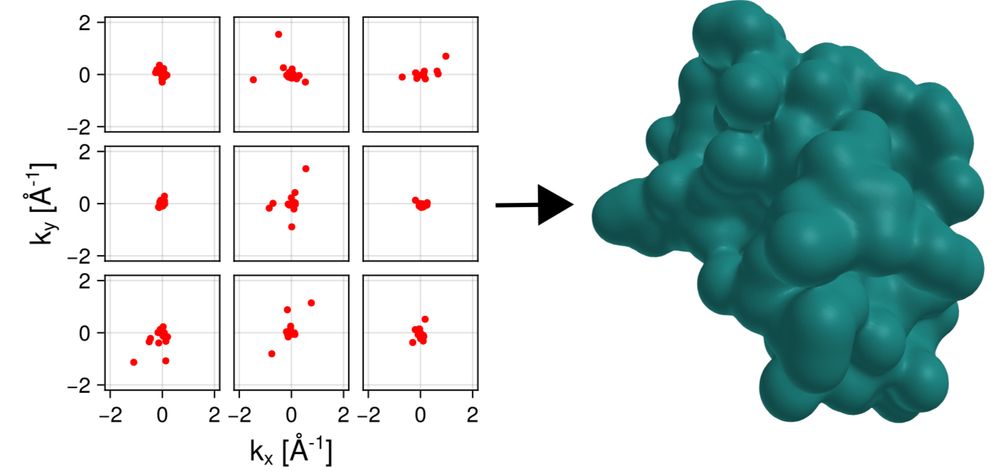
📢 Our new paper in #SciAdv on #Bayesian electron density determination from single-molecule #X-ray scattering is out! 🧐 High resolutions can be achieved from #XFEL exp at low S/N ratios using less than 50 #photons per image! 🔗https://www.science.org/doi/10.1126/sciadv.adp4425 @mpi-nat.bsky.social
10.01.2025 12:10 — 👍 29 🔁 12 💬 0 📌 0
PhD student with @compbiophys | Toronto 🇨🇦 x Göttingen 🇩🇪
Assistant Professor at IRCM, University of Montréal; Adjunct at McGill University. Stem cells, islets & regenerative medicine for T1D. AghazadehLab.com
She/her. Tiohtiá:ke/Montreal. Opinions are my own.
Postdoc @UoD @CeTPD with Alessio Ciulli
Structural & chemical biology in Targeted Protein Degradation
Professor at the Linderstrøm-Lang Centre for Protein Science, University of Copenhagen.
Interests: Protein Quality Control, Genetics, Molecular Chaperones, Degrons.
Spread too thinly. Toronto. Originally from UK. Cancer biology researcher at Sinai Health. Also Prez of the Terry Fox Research Institute.
pharmacist, PhD, researcher, medicinal chemist, drug discovery, neglected diseases, onehealth, Leismania
🔬 Assistant Prof, Pathology @Duke | Director, Clin Micro Lab
🧫 Former Clin Micro Fellow @Memorial Sloan Kettering
👩🏻🔬 Former Postdoc @broadinstitute.org
🎓 PhD @The Rockefeller University
Focus: Diagnostics, AMR, Structural Biology
PhD Student in Integrative structural biology on RNA-binding proteins.
#crystallography #SAXS #NMR #RNA #biophysics #biochemistry #structuralbiology
Molecular biophysicist fascinated by the dynamics of biological systems as captured by time-resolved fluorescence microscopy. Also mountain > sea and cat > dog.
A global community of researchers dedicated to the understanding of proteins.
Asst. Prof. of Chemical Engineering at the University of Toronto| #AI4Science, #MachineLearning and #AI for the Chemical Sciences
Biophysicist and educator
https://www.linkedin.com/in/leonid-brown-92364a4?utm_source=share&utm_campaign=share_via&utm_content=profile&utm_medium=ios_app
Assoc. Prof @polymtl @transmedtech
Cell biophysics, superres microscopy, & curious-methods enthusiast.
We have open positions www.WeissLab.ca
Endurance sports aficionado - Skule and Gopher alum - always eager to learn and up for a chat over a coffee. Single molecule fan - from molecular crystals to protein complexes. Builder of tools and fan of all things imaging
Computational Physicist @ University of Cambridge | physical chemistry | polymers | soft matter
WEB: https://sites.google.com/view/romanstano/
UCSF Biophysics PhD Student. Looking at proteins @fraserlab.bsky.social. Pronouns he/him.
CSO @ Resonate Bio | Vienna - Austria
🧲 NMR Spectroscopy in Drug Discovery and Design | Structural Chemistry | Molecular Recognition
At KITP on the UC Santa Barbara campus, researchers in theoretical physics and allied fields collaborate on questions at the leading edges of science.
www.kitp.ucsb.edu