How do you find newer versions like 2.3?
17.02.2026 16:05 — 👍 0 🔁 0 💬 1 📌 0How do you find newer versions like 2.3?
17.02.2026 16:05 — 👍 0 🔁 0 💬 1 📌 0Thank you!
29.07.2025 12:04 — 👍 0 🔁 0 💬 0 📌 0Can you link the original patient?
25.07.2025 15:03 — 👍 0 🔁 0 💬 1 📌 0Has the ENO and ONE been compared with lower input samples 1-5ng?
27.06.2025 05:58 — 👍 0 🔁 0 💬 0 📌 0Very interesting study, would have been nice to see a SP3 workflow added.
06.06.2025 18:03 — 👍 0 🔁 0 💬 0 📌 0
Excited to share part of my PhD work is now on bioRxiv! We show how LIN28A drives imatinib resistance in CML by reprogramming the kinome. Targeting LIN28A or downstream kinases restores drug sensitivity.
Huge thanks to my amazing collaborators!
#Proteomics #MassSpec #CML #TKIresistance #LIN28A
It’s Canadian, but the Lake Louise Tandem MS is more technological and also good if you like skiing.
20.04.2025 16:26 — 👍 0 🔁 0 💬 1 📌 0Are you using SP3 or S-Trap?
05.04.2025 19:41 — 👍 1 🔁 0 💬 1 📌 0
See if any of the +1 match this list. If so then you have your answer where it is coming from
proteomicsresource.washington.edu/protocols05/...
What are the charge states at the end of the gradient?
05.04.2025 01:56 — 👍 2 🔁 0 💬 1 📌 0Shadowing a postdoc from one of my committee members during my master's.
20.03.2025 13:36 — 👍 2 🔁 0 💬 0 📌 0
I don't have experience with EvoSep. However, they do see it at the end of the gradient in this paper in Fig 5A on Evosep. On our Neo-eclipse setup, I've only ever observed it as the dimer 1021.62 at the end of the gradient when we ramp to 95% B. www.mcponline.org/article/S153...
19.03.2025 12:31 — 👍 0 🔁 0 💬 0 📌 0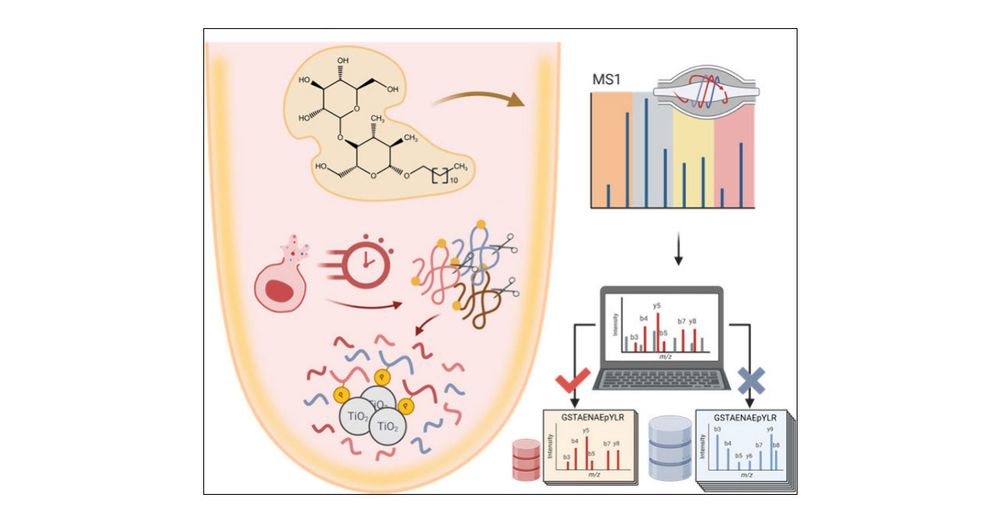
It comes off as a dimer at 1021.62 at the end of gradient. See supplemental figure 3 doi.org/10.1021/acs....
18.03.2025 17:43 — 👍 4 🔁 0 💬 1 📌 0We’ve done a small amount of testing so far with LMNG in digestion buffer and resuspension with DDM, and we have improved recovery. DDM always comes off the end of the gradient as a dimer.
08.03.2025 19:03 — 👍 2 🔁 0 💬 0 📌 0
Also DDM coated tubes improve phosphoproteomics pubs.acs.org/doi/10.1021/...
08.03.2025 16:25 — 👍 2 🔁 0 💬 0 📌 0
This paper compared DDM and other similar surfactants for multiple applications (phospho, AP-MS, EVs and single cell) in digestion and resuspension.
www.mcponline.org/article/S153...
Was this run on a desktop or server?
21.02.2025 01:40 — 👍 0 🔁 0 💬 1 📌 0@resynbio.bsky.social has also been labeled as spam
13.02.2025 13:35 — 👍 1 🔁 0 💬 2 📌 0
MaxQuant YouTube videos are a good starting point. There are also lots of great R scripts.
youtu.be/fdd9MPt2sAU?...

We do DIA without FAIMS on our Eclipse, and it hasn’t given us that much of a boost for our normal workflows. We did see a boost when exploring low inputs below 1ng. Here is a publication of the Olsen lab using FAIMS DIA.
www.mcponline.org/article/S153...

This paper from the Aebersold lab. Depending on the cell type, 0.06 (CMP) to 0.5 (GMP) R^2
11.01.2025 15:10 — 👍 0 🔁 0 💬 0 📌 0Does anyone know how to extract how many MS2s contain particular contaminate fragments? (i.e. the percentage of MS2 with 136.061 m/z Adenine accounting for ppm error) Either with R, python, MSfragger, MaxQuant or Skyline is preferred. #massSOS #proteomics
02.01.2025 16:43 — 👍 0 🔁 0 💬 1 📌 0I'll be back in the lab on January 7th. Can you wait until then?
26.12.2024 22:06 — 👍 0 🔁 0 💬 1 📌 0We've only used Thermos barcode plates. Could you try purchasing a thermos 96-well plate and then making a copy of the barcode or transferring the barcodes to new plates? It looks like I could peel off the barcodes.
26.12.2024 21:13 — 👍 0 🔁 0 💬 1 📌 0HUPO in Toronto Canada
17.12.2024 01:56 — 👍 0 🔁 0 💬 1 📌 0What’s the average peptide length? What’s the cost?
05.12.2024 20:31 — 👍 0 🔁 0 💬 0 📌 0
Alternative enzymes to trypsin give complimentary phosphoproteome. Many amino acids are inaccessible via trypsin.
doi.org/10.1111/febs...
And
doi.org/10.1016/j.ce...
With Ryzen 7950x reaching 32 threads. A threadripper may not be needed for most, but maybe your new TimsTOF ultra will run through samples fast enough to need a threadripper.
12.02.2024 21:16 — 👍 0 🔁 0 💬 0 📌 0I have not compared and don't have access to a threadripper. Though they use the same architecture, I assume IPC would be similar. The main advantage is an increased number of cores and a number of PCI lanes. The number of PCI lanes don't matter much, as we don't need them for multiple GPUs.
12.02.2024 21:12 — 👍 0 🔁 0 💬 0 📌 0I use a Ryzen 2600x with a 1TB 960 Evo M.2 drive and 48 GB RAM. It finishes much faster than any of the more expensive Lab computers with Xeon processors. I keep the task manager open when running FragPipe, MQ and DIA-NN; CPU and disk speed are always limiting factors on all lab computers.
11.02.2024 18:11 — 👍 2 🔁 0 💬 1 📌 0