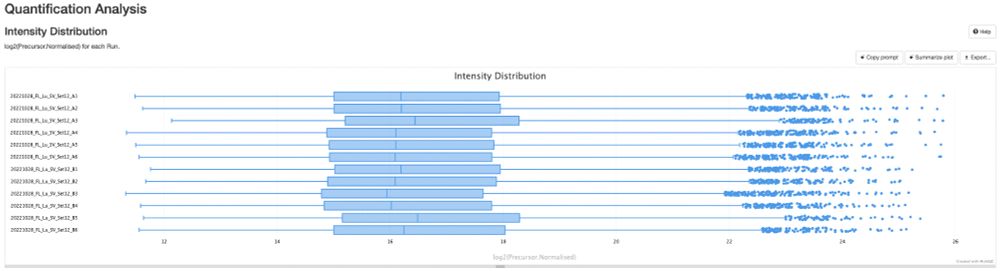
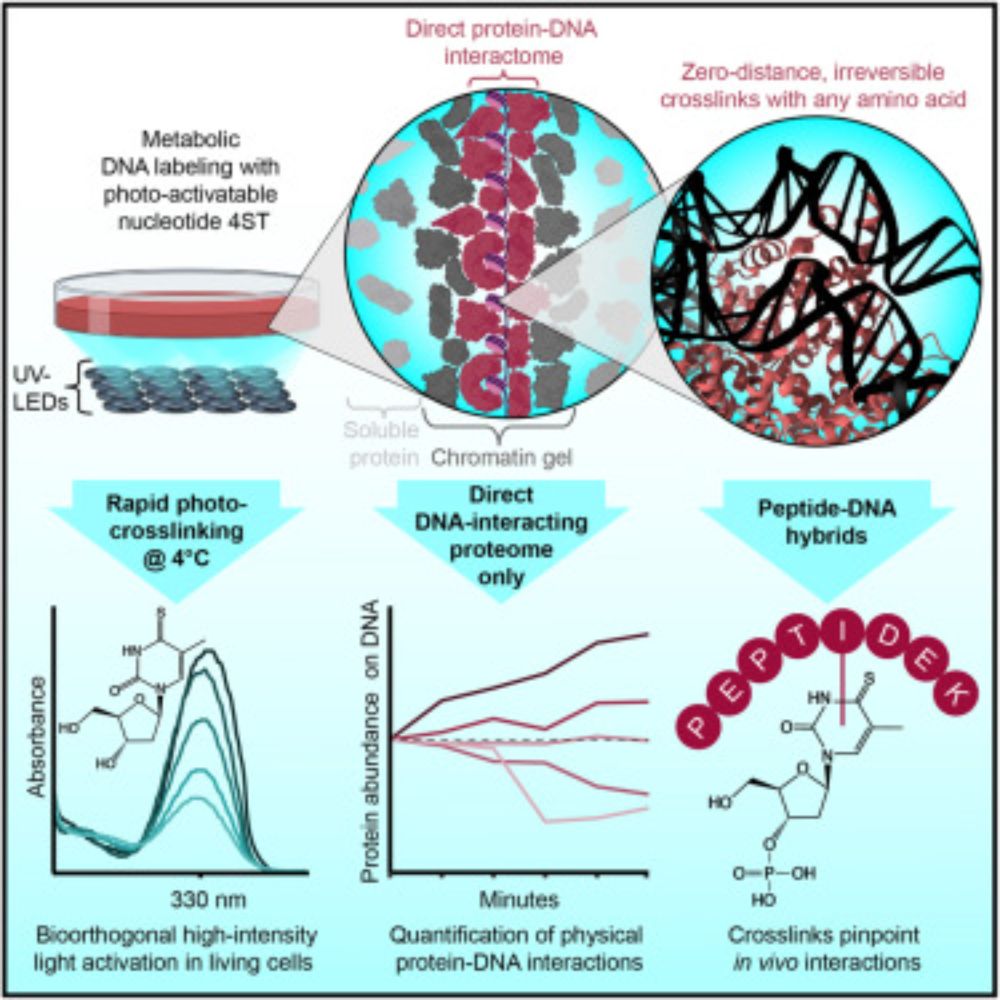
🚀 Excited to share our new preprint: msBayesImpute - A Versatile Framework for Addressing Missing Values in Biomedical Mass Spectrometry Proteomics Data
👉 Improves imputation accuracy, normalization, and differential expression detection
📝https://www.biorxiv.org/content/10.1101/2025.10.02.679746v1
07.10.2025 08:47 — 👍 15 🔁 2 💬 1 📌 1

🥁 Check out our new preprint on OmniPath, the prior knowledge resource for #SystemsBiology, and its brand-new OmniPath Explorer web app! 🥳
📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️
17.09.2025 13:09 — 👍 35 🔁 20 💬 1 📌 0
Abuse of power at universities is a problem in many countries. Unfortunately, the open to read book is just available in German language.
10.09.2025 13:59 — 👍 0 🔁 0 💬 0 📌 0
If you are a #PhD candidate or #student in the fields of #Biochemistry or #ChemBio and would like to go to the #Biochemistry2026 conference in Würzburg, consider applying for the brand new @gdchbiochem.bsky.social Meeting #Award!
en.gdch.de/network-stru...
#ChemSky #ECR #EarlyCareerResearcher
21.08.2025 05:12 — 👍 5 🔁 3 💬 0 📌 0
It's maybe just anecdotal but we moved our QE plus from Freiburg to Würzburg in a new lab. And even after an exploded split flow it's remarkable stable for 10-14 days.
14.08.2025 08:18 — 👍 0 🔁 0 💬 0 📌 0
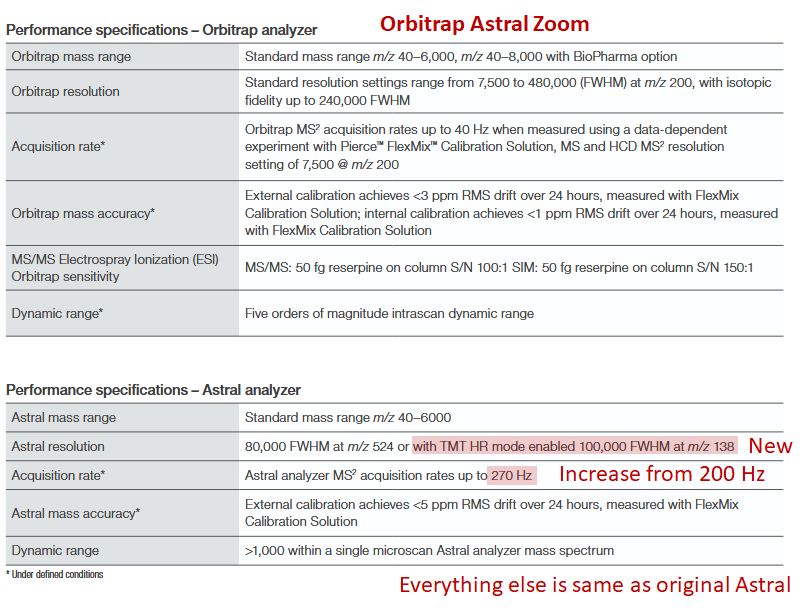
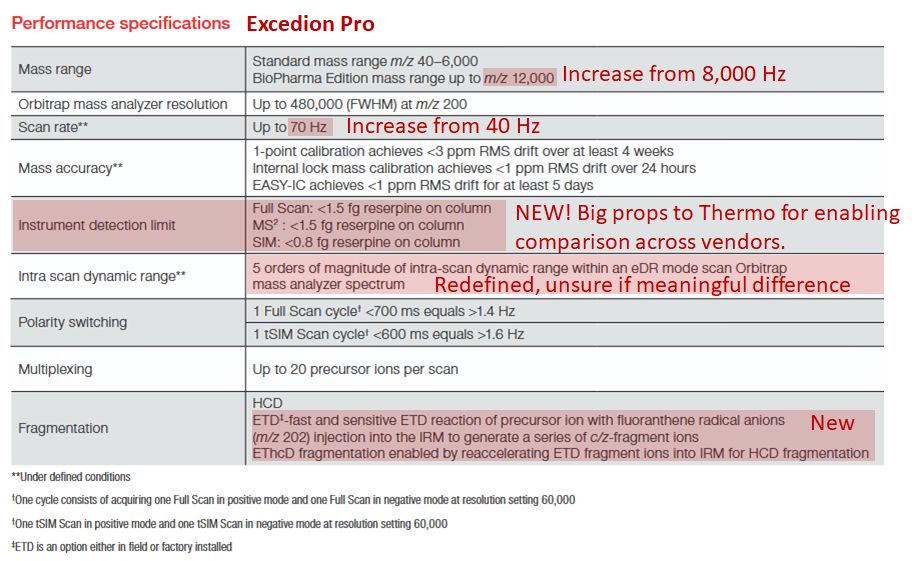
I still find it super curious how different the performance can be. And that their are still gains in precision manufacturing for an already well understood component like the Orbitrap.
13.08.2025 20:37 — 👍 0 🔁 0 💬 0 📌 0
Major part of my PhD thesis is finally online! Thanks to everyone involved 🙏 @walterwchen.bsky.social @rjdlab.bsky.social @tmoeckli.bsky.social @hirakdas.bsky.social and the entire @warscheidlab.bsky.social #peroxisomes #Massspec #proteomics
30.07.2025 10:13 — 👍 12 🔁 3 💬 3 📌 2
On our Orbitrap systems, we normally limit the mass range to 1200m/z in DDA, so I can not give feedback. But still shared your post for better visibility. Hopefully, somebody can help.
31.07.2025 08:07 — 👍 2 🔁 0 💬 0 📌 0
When analysing peptides from in-gel digests by LC-MS/MS, has anyone noticed a 1+ ion of m/z 1342.93 after washing the column with 80% ACN? We are leaving the pump at 80% ACN at 1uL/min for 5 mins after our usual 0-40% ACN gradient. Doesn't correspond to any contaminants. #massspechelp
31.07.2025 07:05 — 👍 3 🔁 1 💬 2 📌 0
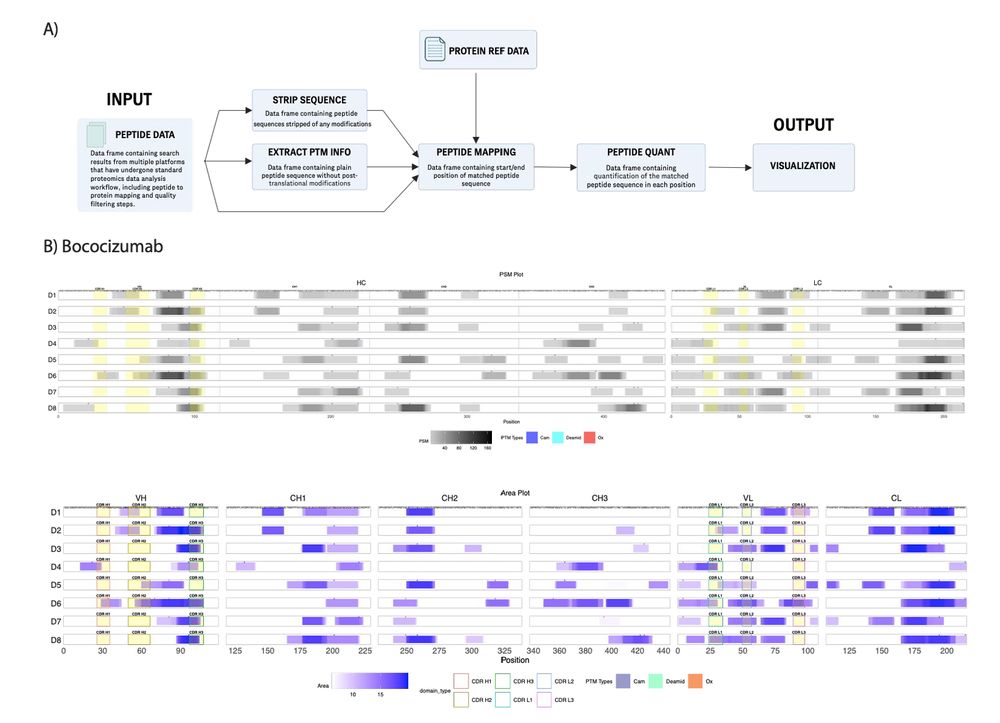
PepMapViz: A Versatile Toolkit for Peptide Mapping, Visualization, and Comparative Exploration academic.oup.com/bio...
---
#proteomics #prot-paper
17.07.2025 14:40 — 👍 11 🔁 5 💬 1 📌 0
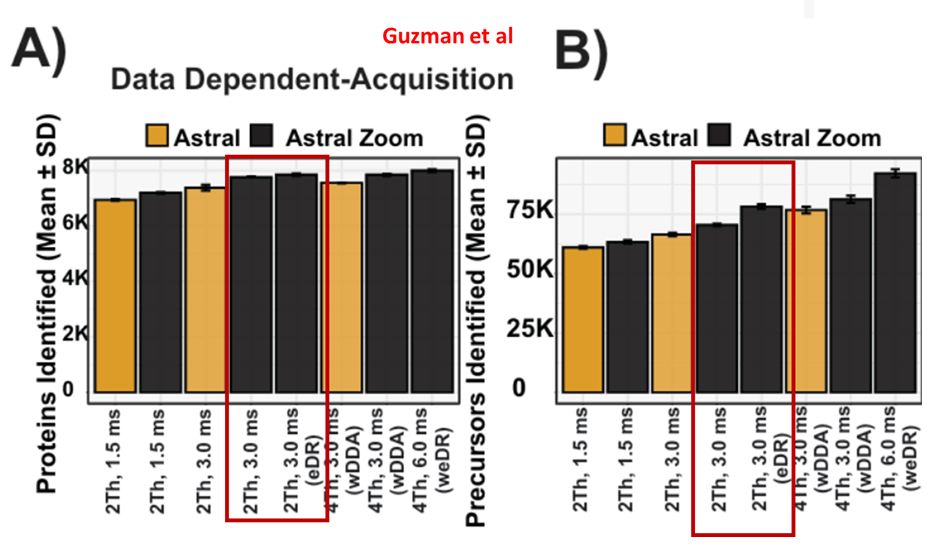
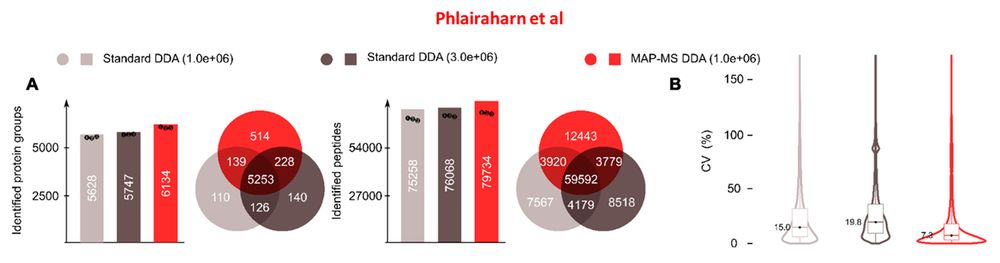
Jesper Olsen Lab (Guzman et al) independently reproduces MAP-MS (eDR) proteome depth and compares Astral Zoom MS to the older model.
"...yielding ~15,000 and ~30,000 additional precursors..."
eDR: www.biorxiv.org/content/10.1...
MAP-MS: www.biorxiv.org/content/10.1...
19.07.2025 09:58 — 👍 20 🔁 4 💬 2 📌 0
As Requested - A Starter Pack of PIs in Biology (or adjacent) fields who are recruiting POSTDOCS! Please don't hesitate to reach out if you wish to be added! go.bsky.app/4gbXXR3
18.07.2025 18:33 — 👍 123 🔁 79 💬 15 📌 3
▶️▶️ To all mass spec enthusiasts: A postdoc position (three years) is available in our international group. Please apply if you are interested in structural MS and in developing novel cross-link ing MS workflows!🧪
Thanks @diegofgar.bsky.social for sharing!
#teammassspec
#AcademicSky
#ChemSky
15.07.2025 13:37 — 👍 15 🔁 15 💬 1 📌 0

Check out our newest paper (tinyurl.com/scp-ms) straight from the lab of @erwinschoof.bsky.social. If you are considering to FAIMS or not to FAIMS and whether wide or narrow window DIA is better, you might find some answers you are looking for. #proteomics
11.07.2025 13:44 — 👍 16 🔁 5 💬 1 📌 0
📣📣It is great to finally welcome @cnicproteomics.bsky.social to Bluesky 🎊 Follow them if you want to learn about the latest development in PTM research applied to cardiovascular diseases! 🫀🧪🧫🖥️🛠️
04.07.2025 17:38 — 👍 3 🔁 1 💬 0 📌 0
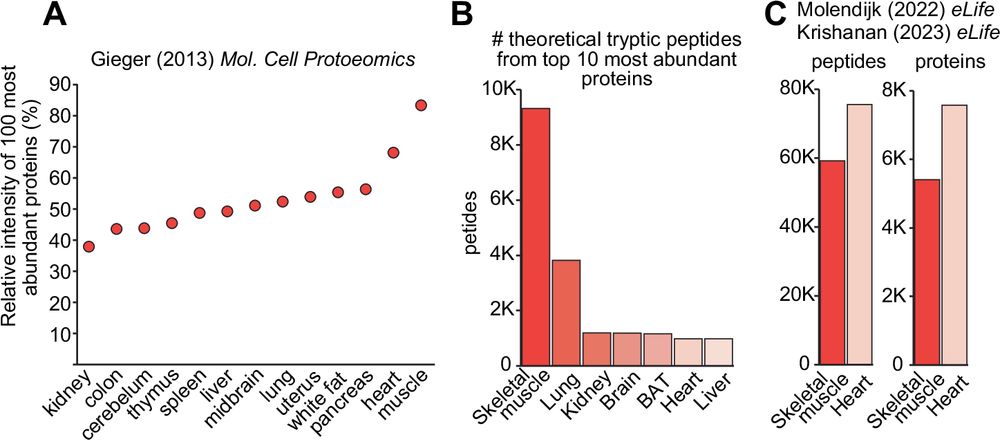
Skeletal muscle proteomics: considerations and opportunities
npj Metabolic Health and Disease - Skeletal muscle proteomics: considerations and opportunities
Are you interested in #proteomics and #skeletalmuscle? It can be a real pain to get deep reproducible coverage. Check out our review with @adeshmukh.bsky.social We analyse some considerations including current limitations and how to potentially overcome them #myoblue @unimelbcmr.bsky.social
03.07.2025 18:17 — 👍 19 🔁 4 💬 0 📌 0
🚀🚀 🚀 Exciting News! 🚀 🚀 🚀
After more than 4.5 fantastic years at @embl.org, I am excited to announce that from 1 August 2025, I’ll be starting my own research group at Physikalisch-Technische Bundesanstalt (PTB) in Braunschweig, Germany — as part of the Biochemistry Department [1/n]
03.07.2025 07:23 — 👍 11 🔁 5 💬 3 📌 0

MaxQuant Summer School 2025 (Utrecht, Netherlands)
16th International Summer School On Computational Mass Spectrometry-Based Proteomics
⏳ Just 1 week left to register for MaxQuant Summer School 2025!
📅 Registration closes: July 7
📍 Utrecht, July 21–25
💻 Hands-on training in MaxQuant, Perseus & mass-spectrometry-based proteomics
🔗 Sign up now: eventbrite.de/e/maxquant-s...
🌐 Details: maxquant.org/summer_school/
#TeamMassSpec
01.07.2025 08:04 — 👍 1 🔁 1 💬 0 📌 0
LinkedIn
This link will take you to a page that’s not on LinkedIn
Anyone interested in using mass spectrometry to identify and quantify peptides and proteins in plasma, tissue and organoid cell systems?
I have a Research Assistant position in my lab at the Institute of Metabolic Science - Metabolic Research Laboratories.
Apply here:
lnkd.in/dCxhS3GN
11.06.2025 15:11 — 👍 7 🔁 5 💬 1 📌 1
Update of our protein outlier caller PROTRIDER. We now handle missing values, a widespread issue for mass spec where missing values are not a random -- and this improves outlier detection on non-missing data! Thumbs up to Daniela and George for the great work.
doi.org/10.1101/2025...
05.06.2025 04:44 — 👍 18 🔁 7 💬 0 📌 0
quantms - Quantitative Mass Spectrometry Analysis
A bioinformatics best-practice analysis pipeline for Quantitative Mass Spectrometry (MS)
🚀 We're launching quantms.org — new hub for all #quantms related projects. Explore its components #pmultiqc & #ibaqpy, and check all reanalysis we're sharing with the community: quantms.org/datasets
Meet the Team and Labs behind the project
👉 quantms.org/about
#OpenSource #Proteomics #ASMS2025
02.06.2025 15:39 — 👍 18 🔁 8 💬 0 📌 0
Very excited that another chapter of my PhD is finally out! 🎉 We compared methods to infer kinase activities from #phosphoproteomics data so check it out if that sounds interesting to you. More details in the thread below 🧵👇🏼
26.05.2025 06:16 — 👍 26 🔁 5 💬 0 📌 0

🧵5 Top Free Alternatives to BioRender for Scientific Illustrations!
These five websites offer free scientific illustrations for biologists. Great for presentations, research papers and other research communication needs.
Save and share the post!
13.05.2025 19:55 — 👍 676 🔁 349 💬 33 📌 6
Group leader at University Hospital Heidelberg
Former Postdoc at EMBL
Bioinformatician, Data scientists Computational mass-spectrometry, multi-omics, and precision oncology. He/Him
https://lu-group-ukhd.github.io/
I’m really good at making vaccines that don’t work. Using glycoproteomics to develop vaccines against flystrike and other parasites for livestock.
Post Doc in Ag and Food at CSIRO. 💉🐑🍬🧬
https://people.csiro.au/k/e/edward-kerr
A Taiwanese in Chicago 👨🔬📯🌈🧋🧋🧋
Proteome explorer, horn player and boba enthusiast!
Research Group: Computational Systems Biochemistry | Max Planck Institute of Biochemistry, Munich 🇩🇪
Proteomics, mass spectrometry, and bioinformatics.
Developers of MaxQuant & Perseus.
Follow our work: https://bsky.app/profile/maxquant.bsky.social
We are the Savitski lab located @ EMBL using and developing proteomics methods for assessing the state of the proteome
Molecular Cell is a Cell Press journal that aims to publish the best papers in molecular biology.
DGMS was founded in 1997 as the successor to the Mass Spectrometry Working Group. It supports and promotes mass spectrometry, a highly precise analytical method for the examination of isolated molecules. dgms.eu
Postdoctoral fellow, Huazhong University of Science and Technology #bioinformatics
Mass spectrometry/Proteomics scientist developing SRM-MS assays to measure Complement proteins in mouse plasma.
Autorin. Politik & Internet. Bücher: Die Daten, die ich rief, Fake Facts, True Facts, Gefährlicher Glaube Podcast: www.denkangebot.org Mastodon: http://chaos.social/@kattascha
Mass spectrometry for high-throughput quantitative proteomics with special interest in posttranslational modifications.
Proteomics at Spanish National Centre for Cardiovascular Research
Old person forgetting everything they ever knew about proteomics at speeds in excess of 270 Hz...
Scientist @UniMelb 🇦🇺 focusing on #proteomics, signaling and metabolism. Loves 🏄♂️⛷️⛰️
Senior Scientist at Evosep, focus on developing workflows for mass spectrometry based biolomecule characterization and proteome profiling. 2 wheels + an engine = fun!
Mass Spec|Proteomics|Chemical Biology
A career network featuring science jobs in academia and industry.
Visit our platform at www.science.hr
Assistant Professor at University of Illinois at Chicago (UIC) in Biological Sciences focused on macromolecular assemblies.
Labormäuserich (Biologe), nur echt mit Mix aus Argumenten und Sarkasmus 😉
🌿🧫🔬🧬
Science, Rumgealber und Musikrätsel