One week left to apply!
08.02.2026 09:04 — 👍 3 🔁 5 💬 0 📌 1Eva Dvorak Tomastikova
@evadt.bsky.social
Scientist working on DNA damage repair in plants Chromatin organization and function group of Aleš Pečinka
@evadt.bsky.social
Scientist working on DNA damage repair in plants Chromatin organization and function group of Aleš Pečinka
One week left to apply!
08.02.2026 09:04 — 👍 3 🔁 5 💬 0 📌 1More info about the topic - contact me on email tomastikova@ueb.cas.cz
08.02.2026 08:10 — 👍 0 🔁 0 💬 0 📌 0
!!!! PhD position!!!!
#SUMO modification in plant DNA damage repair. Join our team @pecinka-grp.bsky.social and work on this cool & still understudied topic. More info ⬇️⬇️⬇️
🌱Send a CV and a brief motivation letter (PDF) to pecinkagroup@gmail.com (subject: Ph.D. Seed development).
🌱The expected start is 1 September 2026.

🚨 PhD position open🚨
Unravel why new polyploids like barley have poor seed viability. Dive into embryo/endosperm development.
Join us! #PhD #PlantBiology #Barley #Polyploidy
A ubiquitin binding protein mediates the crosstalk between PTI and ETI! Check out our new preprint below 👇
07.02.2026 15:38 — 👍 18 🔁 10 💬 0 📌 0
image of a maize inflorescence meristem that is labeled by the plasma membrane marker PIP2-CFP and BAM1D-YFP.
nph.onlinelibrary.wiley.com/share/JAIVZX...
Excited to share my latest work in @newphyt.bsky.social. Here, we used TurboID proximity labeling with CLAVATA receptors FEA3 and BAM1D in maize meristems to better understand the signaling pathways important for meristem regulation. #PlantScience

Coming soon! PhD opening joint with @drjonbaxter.bsky.social at @gdsc-sussex.bsky.social
Keep a look out and get in touch if you are interested in any and all of: Chromosomes, Genome Stability, Chromatin and DNA Topology in Mitosis and Meiosis.
🌱 EPIPLANT is live!
Registration is now open for the EPIPLANT 2026 conference (June 22–25).
We look forward to welcoming you to Nantes, France for an exciting conference on the epigenetics of photosynthetic organisms!
👉 Details & registration: epiplant-2026.sciencesconf.org


I’ll be spending the next four weeks at @agroscope.bsky.social Changins, working on the eccDNA identification in Arabidopsis. Big thanks to @plantepigenetics.ch who agreed to host me and OPJAK Tangenc @iebcas.bsky.social for funding the mobility 😉 🌱🥼🔬
03.02.2026 14:29 — 👍 5 🔁 2 💬 1 📌 0
A zz plant in a brown plot. The plant has waxy lush green leaves.
Today I learned that the common houseplant Zamioculcas (the zz plant) is monocot w/ pinnately compound leaves. So called leaves are in fact leaflets. Unlike most species w/ compound leaves, the leaflets of zz plants are highly regenerative, from which whole plants can be readily regenerated.
28.01.2026 01:32 — 👍 27 🔁 5 💬 0 📌 1The HeatDDR doctoral network training school in full swing @iebcas.bsky.social @pecinka-grp.bsky.social @zeiss-microscopy.bsky.social #microscopy #DNAdamage
28.01.2026 07:15 — 👍 3 🔁 3 💬 0 📌 0
📢 Three new #bioRxiv preprints from our team on holocentric chromosomes.
Together, they connect centromere repeat evolution, karyotype dynamics, and meiotic recombination outcomes, revealing how holocentric genomes evolve and function. 🧬👇



We are hosting the training school of the HeatDDR PhD program over this week at the Center for Structural and Functional Plant Genomics of the Institute of Experimental Botany, Olomouc! 🧬
Doctoral students of the network from all over Europe will learn cutting edge techniques related to DNA damage!
Excited to share our latest paper showing how heat stress halts DNA replication and mitosis in barley roots! 🌡️🌱 #PlantScience #HeatStress #Barley
www.frontiersin.org/journals/pla...


Starting off the new year with a kick-off meeting of a new collaborative grant with
Jan Paleček from Brno. We're excited to unravel how new candidates of DNA damage repair work their magic in Arabidopsis! ☘️ 🧪 🧬
P.S. Good brainstorming needs good pizza🍕


🌟 Wrapping up the year with Christmas sweets and mulled wine at our final lab meeting before Christmas! 🎄#HappyHolidays
20.12.2025 13:37 — 👍 3 🔁 1 💬 0 📌 0


❄️Proper Winter is here!!! ❄️
So we had to try out the snow. ⛷️🏂




We're thrilled to welcome Manuel Escalona Sendín from Spain to our group! Manuel will be diving into the effects of temperature stress on DNA damage in barley. #HeatDDR
He's already sampled Czech cuisine and beer 🍻—and even a snowballs fight! ❄️ #phdlife
Starting on September, 2026, our doctoral student will be a part of the Marie Sklodowska-Curie Actions Doctoral Network Consortium AGILE, involving leading European Labs in advanced microscopy of plant chromosomes.
🧪☘️🧬

We're thrilled to announce our new PhD position in the AGILE Doctoral Network on nuclear lamina's role in the maintenance of Rabl configuration in barley!🔬🌾🧬
More info on the position : olomouc.ueb.cas.cz/en/lamina-in...

It's finally out!...
Using single-cell and spatial transcriptome data, and data imputation, we created BARVISTA, a website able to perform almost genome-wide information on gene expression profiles at single-cell resolution in barley spikes!
www.nature.com/articles/s41...

Great response so far for the DNA Repair Meeting in April 2026 ! 🧬
A few poster spots are still available.
✨ Early-bird registration extended to February 28th
✨ New invited speakers added: check the site: dnarepairmeeting-egmond2026.com
Don’t miss this outstanding meeting!

paper is out now in Nature Plants, here is a link: Imputation integrates single-cell and spatial gene expression data to resolve transcriptional networks in barley shoot meristem development. Nat. Plants (2026). doi.org/10.1038/s414...
08.01.2026 00:49 — 👍 60 🔁 33 💬 0 📌 1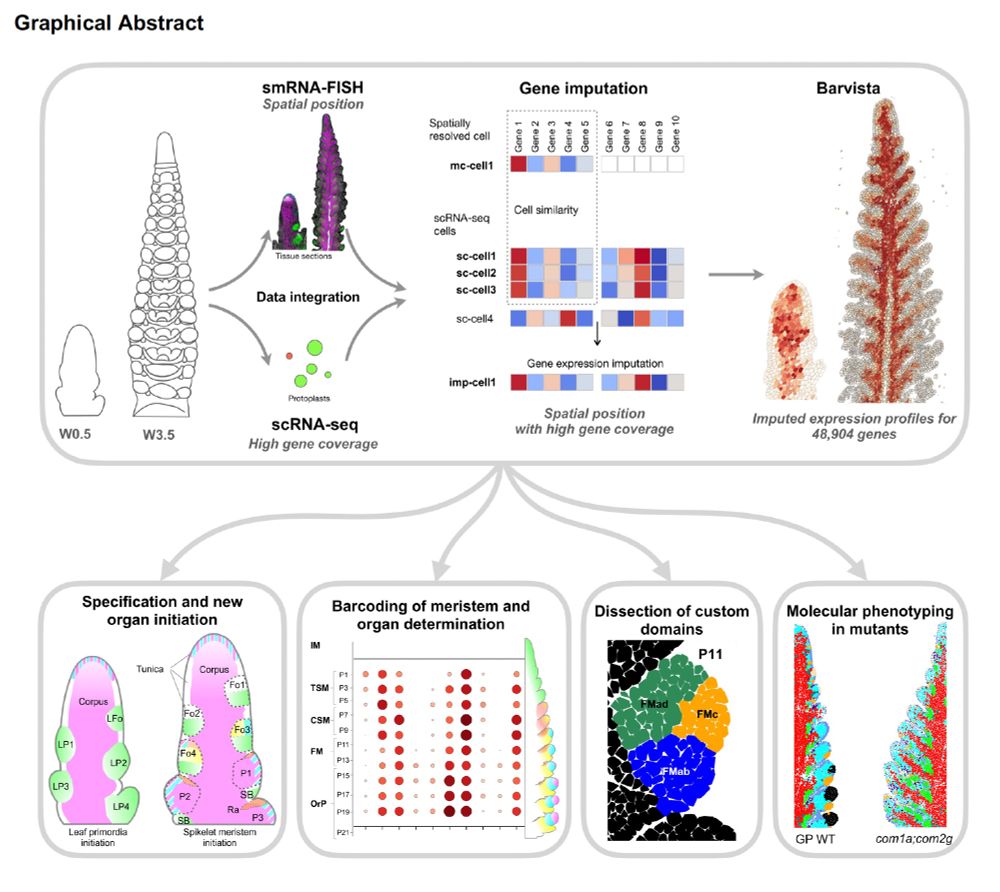
Meristems shape plant architectures, and grasses generate a complex array of them. To characterise barley vegetative SAMs and spike development, we used single cell and spatial transcriptome data and integrated them in a new database, BARVISTA. A click on a cell now.....
tinyurl.com/38c3mf5d

🚀 Qlife Winter School 2026: Quantitative Approaches to DNA Damage and Repair! Hands-on workshops for Master's, PhD, postdocs & junior scientists. 🗓 Apply by Jan 19, 2026! #QlifeWinterSchool #QuantitativeBiology #LifeScienceTraining
06.01.2026 10:34 — 👍 1 🔁 2 💬 0 📌 0
Identification d’un mécanisme global de répression de la transcription chez la plante modèle Arabidopsis thaliana
16.12.2025 21:58 — 👍 1 🔁 2 💬 0 📌 0
Fig. 1 (shortened, full legend in paper): Complexity of gynoecium development shown with confocal microscopy imaging. (A) Meristematic stages of floral development in which the inflorescence meristem (IM) is observed; from stage 3, the sepal primordia (asterisks) are observed; in stage 4, the developing sepals (asterisks) partially cover the floral meristem (FM); and in stage 5, the FM is completely covered by the sepals. (B) Stage 6 in which the gynoecium is established with the appearance of the primordium of the gynoecium. (C) In stage 7, the middle and lateral domains can be differentiated (marked by dotted lines). (D–H) Longitudinal view of the gynoecium from stage 8 to 12. At stages 8 and 9, the gynoecium grows as a hollow tube and the valves and young replum can be seen. (F and G) Longitudinal view of stage 10 and 11 gynoecia showing the replum and valves. (H) Mature stage 12 gynoecium showing the stigma, style, and ovary, the latter formed by the valves and replum.
🌸 Flowering Newsletter Review 🌸
López-Gómez et al. review the role of protein–protein interactions in flower and fruit development and discuss the latest techniques to study them.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪 @defolter-lab.bsky.social

Plant Science Research Weekly -- Review: A complete guide to metabolomics (Plant Physiol.) (Summary by Mary Williams @PlantTeaching.bsky.social) buff.ly/ocoixG3
#PlantaePSRW

A Single-Cell-Resolution Spatial Transcriptomic Atlas Decodes Wheat Spike Development and Yield Potential #resource #MolecularPlant cell.com/molecular-pl...
25.12.2025 08:07 — 👍 8 🔁 6 💬 0 📌 0