Weta workshop?
11.08.2025 09:11 — 👍 0 🔁 0 💬 1 📌 0Niko Klink
@nikolasklink.bsky.social
Chemical Biology & drug discovery @Max Planck Dortmund | All things Ubiquitin and bifunctionals @Gersch Lab.
@nikolasklink.bsky.social
Chemical Biology & drug discovery @Max Planck Dortmund | All things Ubiquitin and bifunctionals @Gersch Lab.
Weta workshop?
11.08.2025 09:11 — 👍 0 🔁 0 💬 1 📌 0Very happy to see our contribution to a better understanding of Hsp70/90 #chaperones online! Congrats and a big thank you to all co-authors, collaborators and reviewers! #proteostasis @poepsel-lab.bsky.social @crc1430.bsky.social @unidue-zmb.bsky.social
15.07.2025 06:06 — 👍 32 🔁 17 💬 1 📌 1The degrader is much much worse in cellular permeability than the inhibitor (we tested), but is just so potent at forming a stable ternary complex and inducing degradation that we just need less for full degradation!
10.07.2025 06:19 — 👍 0 🔁 0 💬 0 📌 0Thanks alot, much appreciated, and thanks for the feedback! Would you like to see inhibitor go down or degrader go up with the concentration? Equimolarity is a bit iffy to compare here, since they have such drastically different membrane permeability
09.07.2025 06:43 — 👍 0 🔁 0 💬 0 📌 0And wishing you a full and speedy recovery!
06.07.2025 09:45 — 👍 1 🔁 0 💬 1 📌 0Ah now I understand your question correctly, i thought you meant something different! Sorry to hear that. I dont know enough about long term treatment of Kras inhibitors to answer that question truthfully, sorry!
06.07.2025 09:44 — 👍 1 🔁 0 💬 1 📌 0Yes, it will very much hinder degradation! The inhibitor has higher cell membrane permeability and will outcompete the Degrader binding site
06.07.2025 09:30 — 👍 2 🔁 0 💬 1 📌 0Thank you! By becoming available later you mean Treating cells with inhibitor first then adding degrader at later time points?
06.07.2025 09:25 — 👍 1 🔁 0 💬 1 📌 0Lastly, a huge thank you to my supervisor Dr. @maltegersch.bsky.social for his guidance and support, as well as the entire Gersch team.
I am very glad to share this story, but even more excited for whats to come in the (near) future, so stay tuned, we are just getting started!
#TPD #PROTAC #DUB
Big thanks also to Dr. Bikash Adhikari and Prof. Elmar Wolf from @wolflab23.bsky.social, as well as Dr. @martinschwalm.bsky.social for bringing their expertise to this project, @imprs-lm.bsky.social from the MPI Dortmund for the excellent support and everyone else who contributed to the prokect!
05.07.2025 09:51 — 👍 1 🔁 0 💬 1 📌 0This work would not have been possible without my amazing co-first authors Dr. Sebastian Urban and Dr. Med. Johanna Seier and everyone in this highly interdisciplinary team from the @crc1430.bsky.social and beyond.
05.07.2025 09:45 — 👍 0 🔁 0 💬 1 📌 0
Taking these now to our Model Systems (PDAC and melanoma), not only shows highly selective degradation of USP7 by our PROTACs (criteria for matched pair fulfilled!), but also striking differences between inhibitor and degrader treated cells. These differences also occur in many phenotypic assays!
05.07.2025 09:42 — 👍 1 🔁 0 💬 1 📌 0
After making a bunch of degraders (most not shown here), we arrive at NK250 and NK266 ("Kurt Cubane"), two highly potent USP7 degraders in HiBiT and immune-based assays. NK250, is a faster degrader which we could correlate to its ability to form a stable ternary complex between USP7 and VHL.
05.07.2025 09:41 — 👍 0 🔁 0 💬 1 📌 0
We introduce NK192, a potent USP7 inhibitor of the widely used hydroxypiperidine scaffold. Converting it through the available exit vector to a biotin probe showed proteome-wide highly selective binding to USP7, fulfilling our criteria for the inhibitor side of the matched chemical pairs.
05.07.2025 09:39 — 👍 0 🔁 0 💬 1 📌 0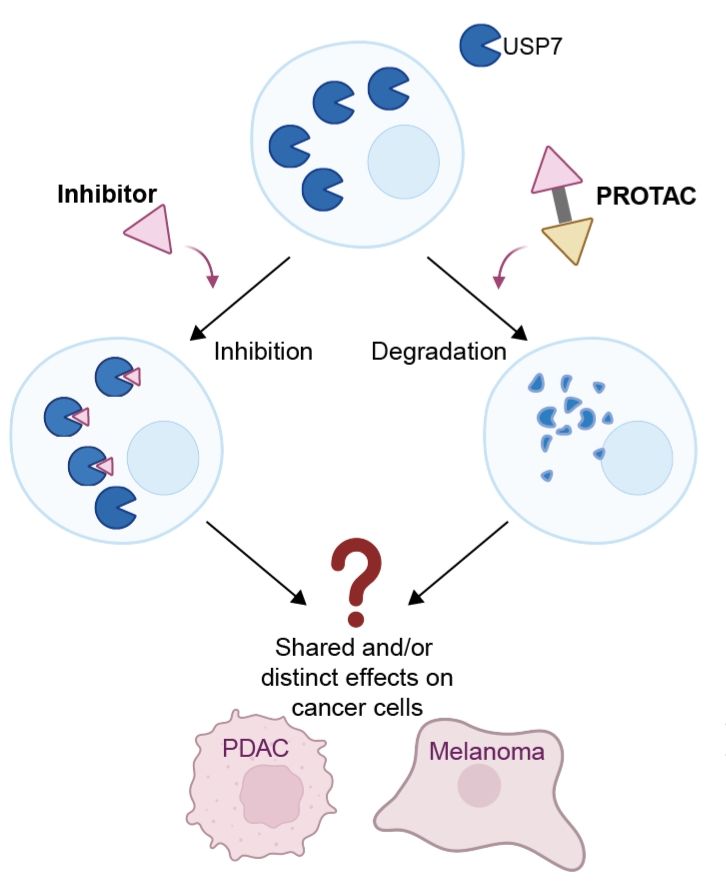
To enable their systematic discovery, we addressed this urgent need with matched chemical pairs of DUB inhibitor and #degraders by focusing on USP7 as a case study. By using potent and selective modulators, we aimed to dissect phenotypes induced by inhibitors and degraders of USP7 in solid cancers.
05.07.2025 09:36 — 👍 0 🔁 0 💬 1 📌 0#Deubiquitinases (DUBs), key regulators of ubiquitin signaling, have been increasingly recognized for functions beyond rescuing substrate proteins from proteasomal degradation. Due to a lack of selective chemical tools within the DUB space, finding these functions has proven to be very challenging.
05.07.2025 09:34 — 👍 0 🔁 0 💬 1 📌 0
Really excited to share the first chapter of my PhD as a preprint titled: Targeted degradation of USP7 in solid cancer cells reveals disparate effects of deubiquitinase inhibition vs. acute protein depletion: www.biorxiv.org/content/10.1.... If that sounds interesting to you, small tweetorial below👇
05.07.2025 09:33 — 👍 15 🔁 2 💬 2 📌 1We hope that our work paves the way for the next generation of molecules targeting USP30 and fighting neurodegenerative diseases, and hope that this strategy will soon be applied to other DUBs!
05.05.2025 18:50 — 👍 2 🔁 0 💬 0 📌 0Huge shoutout to Nafizul Kazi for this incredible tour de force on tireless protein engineering and thorough biochemical assay characterizations, @gallantkai.bsky.social and Gian Marvin Kipka for the beautiful cellular work and @maltegersch.bsky.social for amazing guidance and supervision.
05.05.2025 18:48 — 👍 3 🔁 0 💬 1 📌 0
Very excited to share that our work on Parkinson's target USP30 has been published in Nature Structural & Molecular Biology! By combining soluble DUB-domains onto USP30, we for the first time achieved co-crystallization with a small molecule (NK036) without Nanobodies! www.nature.com/articles/s41...
05.05.2025 18:42 — 👍 11 🔁 0 💬 1 📌 1Absolutely fantastic work, congrats to the whole team!
25.03.2025 05:51 — 👍 1 🔁 0 💬 1 📌 0
Wow. This is really stunning . www.science.org/doi/10.1126/...
21.03.2025 18:02 — 👍 55 🔁 8 💬 1 📌 2
I'm super excited to share our study in collaboration with @jnpruneda.bsky.social, published today in @embojournal.org! We show that PARPs are modified with a non-canonical ester-linked mono-ADP-ribosylated ubiquitin species for the first time in cells: www.embopress.org/doi/full/10....
25.02.2025 17:52 — 👍 50 🔁 20 💬 4 📌 2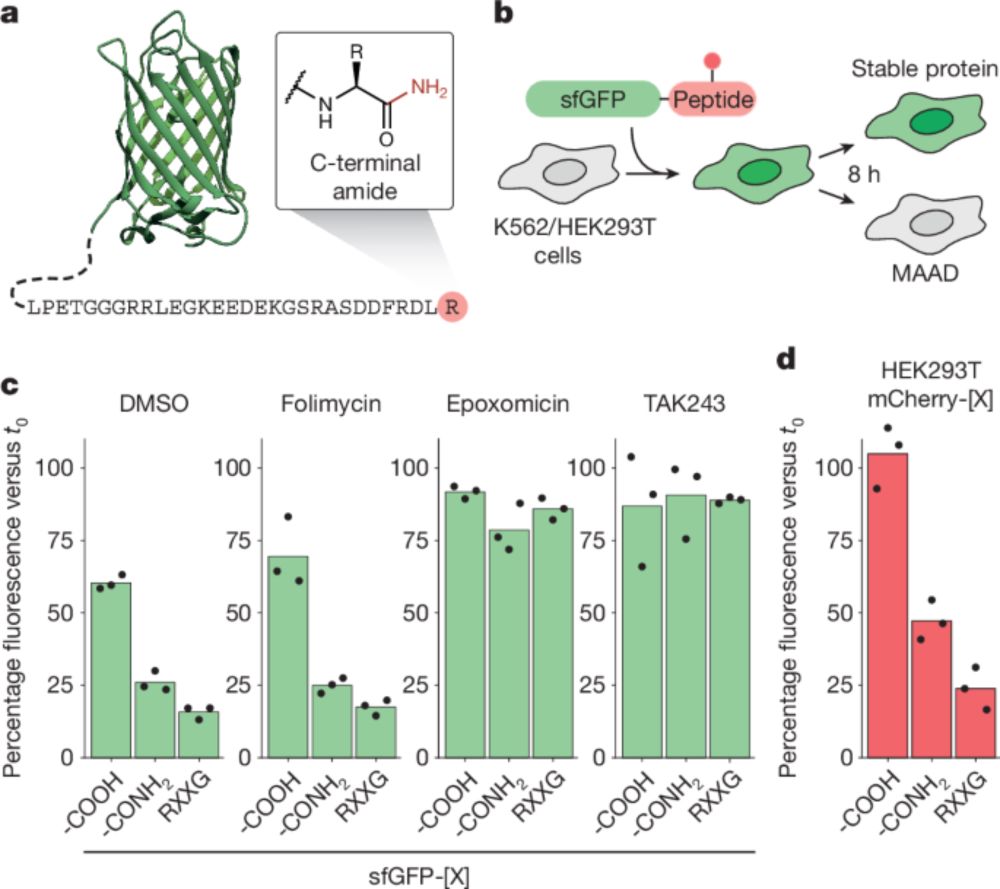
🎉Super excited to share our story on how the substrate receptor FBXO31 functions as a quality control factor by recognizing amides. This has been an amazing collaboration between Bode lab and @jcornlab.bsky.social. Special shutout goes to @matthiasmuhar.bsky.social www.nature.com/articles/s41...
29.01.2025 16:29 — 👍 85 🔁 35 💬 3 📌 4I dont recall where I read it, but dont These have rather Poor affinity for CRBN compared to the usual suspects?
23.12.2024 14:41 — 👍 0 🔁 0 💬 0 📌 0Congrats Georg, fantastic news and well deserved!
03.12.2024 12:14 — 👍 1 🔁 0 💬 1 📌 0A fantastic story from our lab about finding novel activity in two "inactive" DUBs!
25.11.2024 19:10 — 👍 7 🔁 0 💬 0 📌 0go.bsky.app/3VPQofn
Great way to get started here!