High-throughput mapping of 6,888 RAD51D variants identifies distinct biochemical functions needed for homologous recombination and olaparib response https://www.biorxiv.org/content/10.64898/2026.01.11.698865v1
12.01.2026 02:47 — 👍 0 🔁 1 💬 0 📌 0
And a poster today (9259W) from talented undergrad researcher Ellie Bloss. Then, on Friday, two coding MAVEs for the price of one, from amazing postdoc Seba Vishnopolska (poster 5039F)
15.10.2025 14:20 — 👍 3 🔁 0 💬 0 📌 0

excited to be at #ASHG25! Lab has brought a series of functional screens - talks tomorrow from postdoc Adelaide Tovar @adel-aide.bsky.social (945am) and clinical asst prof Tony Scott (215pm) showing a pair of high-throughput functional screens.
15.10.2025 14:17 — 👍 4 🔁 2 💬 0 📌 1
Asst Professor | U-M Careers
The Department of Human Genetics at the University of Michigan is looking for a new faculty colleague at the rank of Assistant Professor. Please share!
careers.umich.edu/job_detail/2...
25.09.2025 18:26 — 👍 12 🔁 13 💬 0 📌 0
Delighted for this to be published at AJHG @ajhgnews.bsky.social ! Updates vs the preprint include new comparisons to AlphaMissense and other predictors. We hope this map will help improve clinical variant interpretation in MUTYH! #VUS #VariantEffects #ColorectalCancer www.cell.com/ajhg/fulltex...
06.08.2025 18:40 — 👍 3 🔁 2 💬 0 📌 0
They don't care that republicans still get cancer and other diseases and fall on hard times and have special-needs kids. This was the deal. "We will hurt the people you hate even if it hurts you" and half the country signed up in blood
30.06.2025 00:49 — 👍 117 🔁 20 💬 4 📌 2


We hope this serves as a useful resource to assist with rare variant interpretation, and believe our approach can be extended to other DNA repair factors where the #VUS burden challenges the actionability of genetic testing. 5/5
07.03.2025 19:27 — 👍 0 🔁 0 💬 0 📌 0
We examined individual-level clinical phenotypes of MUTYH biallelic individuals, in collaboration with a clinical lab (Ambry Genetics). Individuals with a pathogenic+missnse VUS had much higher polyposis risk -- when their VUS scored as abnormal by our assay 4/5
07.03.2025 19:27 — 👍 0 🔁 0 💬 1 📌 0
Our map perfectly separates known pathogenic and benign variants from ClinVar, and based upon clinical calibration, it provides ‘strong’ evidence to resolve the >1000 standing clinical missense VUS 3/5
07.03.2025 19:27 — 👍 0 🔁 0 💬 1 📌 0
To measure MUTYH variant function, we leveraged a reporter containing a mispair of 8oxoguanine w/ adenine, the substrate that MUTYH recognizes. AFAIK, this is the first MAVE coupled to a direct readout of repair at an DNA oxidative damage lesion. 2/5
07.03.2025 19:27 — 👍 0 🔁 0 💬 1 📌 0
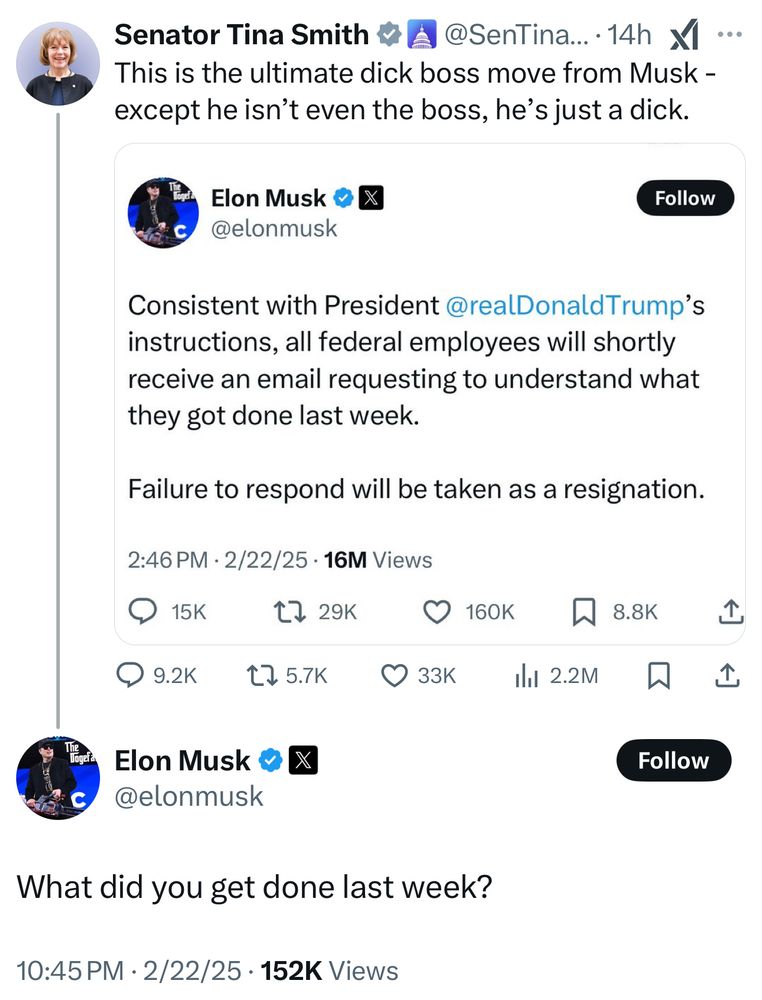
Senator Tina Smith said: This is the ultimate dick boss move from Musk - except he isn’t even the boss, he’s just a dick.
That was in response to Musk saying “Consistent with President @realDonaldTrump's instructions, all federal employees will shortly receive an email requesting to understand what they got done last week. Failure to respond will be taken as a resignation.”
Musk responded @SenTinaSmith “What did you get done last week?”
Elon, I hate to break it to you but you aren’t my boss. I answer to the people of Minnesota.
But since you bring it up, I spent last week fighting to stop tax breaks for billionaires like you, paid for by defunding health care for moms and babies
23.02.2025 15:26 — 👍 22210 🔁 4186 💬 635 📌 314

In short: SpliceAI and Pangolin generally performed well. Splicing effects are harder to predict in exons vs introns. Gene model annotation really matters! We think the places where they disagree will be interesting targets for future MPSAs. 3/3
21.12.2023 15:57 — 👍 0 🔁 0 💬 0 📌 0
This leverages MPSAs from our group and others. Cathy led several of these, including one published this past summer in WT1, a transcription factor where alt splicing is disrupted in Mendelian forms of nephrotic syndrome www.kireports.org/article/S246... 2/3
21.12.2023 15:57 — 👍 0 🔁 0 💬 1 📌 0

Last paper from the group for 2023, now out at Genome Biology! Led by recent bioinformatics grad Cathy Smith, we used #VariantEffect maps from massively parallel splicing assays (MPSA) to test bioinformatic splicing effect predictors genomebiology.biomedcentral.com/articles/10.... 1/3
21.12.2023 15:56 — 👍 2 🔁 0 💬 1 📌 0
Finally, also on Thursday, PB1111 from talented undergrad Kirsten Nishino (also w/ Steve Parker & Adelaide Tovar) examines how environment effects modify transcriptional response using this modular MPRA system 5/5
01.11.2023 18:53 — 👍 0 🔁 0 💬 0 📌 0
On Sat, PB1089 from postdoc Adelaide Tovar (& Steve Parker lab) uses a modular MPRA to interrogate enhancer-promoter context and cell type influences on regulatory action 4/5
01.11.2023 18:53 — 👍 0 🔁 0 💬 1 📌 0
Also, PB1030 from 2xDr Anthony Scott and colleagues at Ambry Genetics shows how we've calibrated another deep mutational scan of another #LynchSyndrome gene, MLH1, for use in clinical variant interpretation 3/5
01.11.2023 18:53 — 👍 0 🔁 0 💬 1 📌 0
Check out these posters on Thurs too! In PB1006, we present a really cool (imho) DMS strategy from postdoc Shelby Hemker which goes beyond drug or viability selections to read out function of DNA repair factor variants 2/5
01.11.2023 18:53 — 👍 0 🔁 0 💬 1 📌 0
So many great talks and posters to see at #ASHG23 #ASHG2023! Here are a few from my group. Thurs 11:15AM (session 23), postdoc Seba Vishnopolska will present a deep mutational scan of a key #LynchSyndrome factor PMS2. 1/5
01.11.2023 18:53 — 👍 4 🔁 0 💬 1 📌 0
Founder & reigning monarch at TPM. Lapsed historian. Hand tool woodworker. Jew.
Functional genomicist & Assistant Professor at The University of Texas Medical Branch at Galveston. https://www.ward-lab.org/
Same content, different website! Human evolutionary genomics, functional genomics and archaic hominins. Group leader in Human Genomics at SVI in Melbourne/Naarm, Australia. Durian evangelist. Sometimes I go to Estonia.
🧬Founded in 1948, the American Society of Human Genetics (ASHG) is the primary professional membership organization for #humangenetics specialists worldwide.
www.ashg.org
Professor | Molecular Medicine | Halle University
Pediatric Hematologist/Oncologist, Geneticist, Stem Cell Biologist
bloodgenes.org
Ex NY Times, now author of Substack Paul Krugman. Nobel laureate and, according to Donald Trump, "Deranged BUM"
Bronx boy. Cubs fan. Dad, husband, writer, podcaster and cable news host.
The Sirens’ Call: How Attention Became the World’s Most Endangered Resource out now.
https://sirenscallbook.com/
Senior Investigator @ Altius Institute for Biomedical Sciences. Research: High-resolution mapping of chromatin structure & function. Fun: Mountain shenanigans and skiing turns all year. Seattle, USA/Patagonia Chilena (🇺🇸🇨🇱). http://vierstra.org
Retweets of alternative splicing tweets.
Account managed by @m1quelag.bsky.social
Genomics, technology and human genetics @University of Washington. Working to create an atlas of variant effects and resolve VUS.
Advancing the promise of the Human Genome Project by interpreting the landscape of human genetic variation.
https://www.varianteffect.org
https://www.linkedin.com/company/atlas-of-variant-effects-alliance
Assistant Professor of Molecular Genetics at the University of Toronto's Donnelly Centre
Research Assistant Professor with Gemma Carvill (@CarvillLab on Twitter). Focus: epilepsy genetics. Twin. Ace cat dad. Occasional writer. I am only an egg. He/him.
Associate Professor at TGen.org. Interested in genmoics, single cell and spatial transcriptomics, lung disease, and oftentimes fishing. banovichlab.org
Asst Prof of Biology at Gonzaga University. Pharmacogenomics, yeast genetics and evolution, STEM education. UW Genome Sci and Harvard BBS alum.
Genome evolution, podcasts, toast, and popcorn
genetics, genomics, and gossip • NHGRI #NIHMOSAIC K99/R00 fellow at Michigan, formerly at MIT/UNC
https://adelaidetovar.com
Bloomberg Distinguished Professor at Johns Hopkins University. http://schatz-lab.org











