Ultra-low-field brain MRI morphometry: T-RT reliability and correspondence to high-field MRI | doi.org/10.1162/IMAG... | w/ !! FREE DATA !! T1w & T2w scans of 23 healthy participants from 3T GE Premier & 2x 64mT Hyperfine Swoop in many resolutions, i.e. 390 scans! openneuro.org/datasets/ds0... [1/n]
29.09.2025 14:49 — 👍 5 🔁 3 💬 1 📌 0
This was a great collaboration with @joachimdejonghe.bsky.social and @chloeterwagne.bsky.social
15.10.2025 14:41 — 👍 2 🔁 2 💬 0 📌 0
We're recruiting early career Group Leaders this autumn! I cannot think of a better place to build a lab. Come join us! 👇
09.10.2025 14:59 — 👍 5 🔁 1 💬 0 📌 0
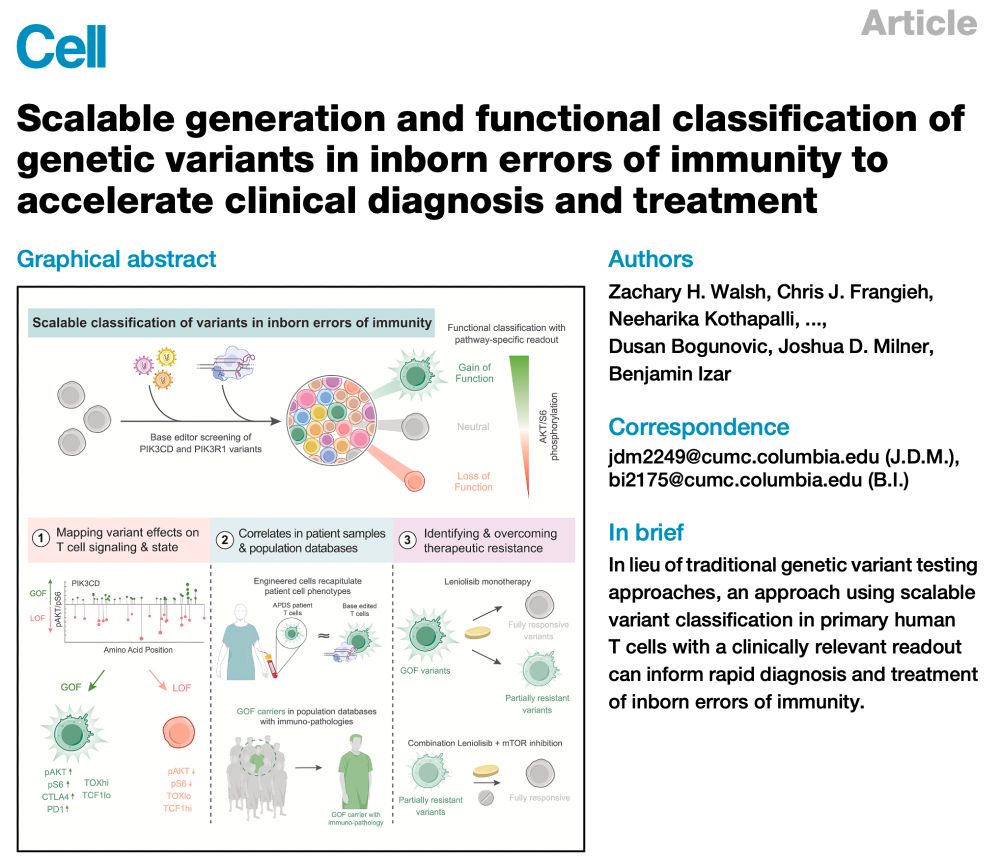
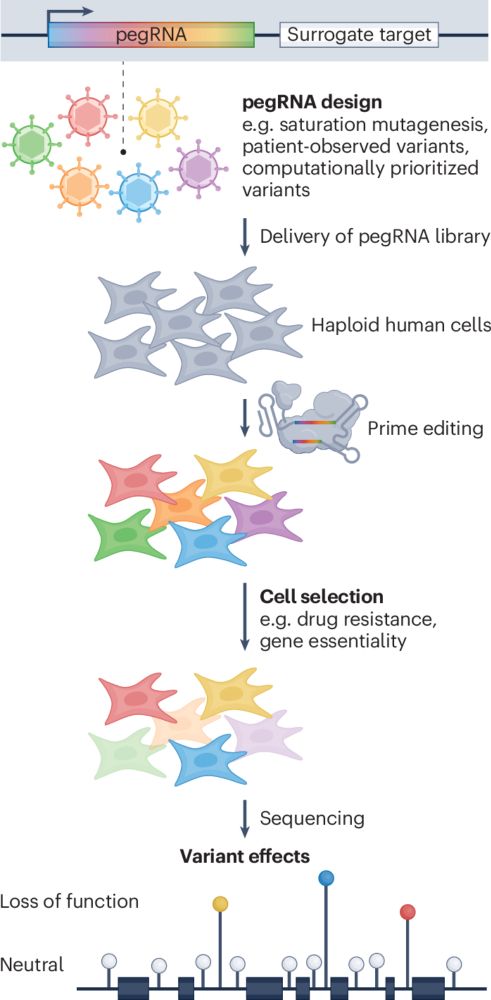
Thrilled to re-share our tweetorial on Bluesky: now out in @cp-cell.bsky.social (🔗 tinyurl.com/3a55tsky) - we present a framework to accelerate variant classification, diagnosis & treatment of inborn errors of immunity. A dream MD/PhD project, which has already led to the treatment of a patient.
02.07.2025 16:42 — 👍 20 🔁 8 💬 2 📌 0
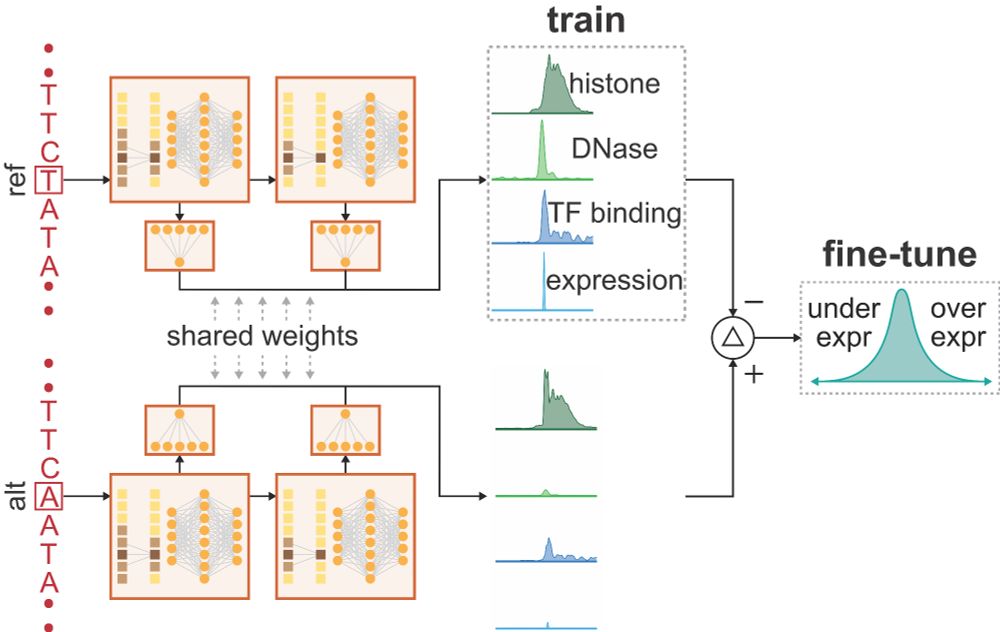
We're thrilled to introduce PromoterAI — a tool for accurately identifying promoter variants that impact gene expression. 🧵 (1/)
29.05.2025 18:29 — 👍 59 🔁 28 💬 1 📌 2
We're quite excited about this story as it showcases the power of SGE to dissect non-coding genes and to uncover new disease associations and diagnoses. This has, indeed, been an excellent collaboration...
11.04.2025 16:14 — 👍 23 🔁 8 💬 1 📌 0
Hats off to @joachimdejonghe.bsky.social for overcoming some major hurdles to pull this off. And many, many thanks to @nickywhiffin.bsky.social and co for taking SGE data to new heights. It's been amazing to see how fast things can move once we know precisely which variants matter.
11.04.2025 16:14 — 👍 4 🔁 1 💬 1 📌 0
Close all tabs and read this. Start with the preceding Nature paper. Trust me. This is cooler than liquid helium.
11.04.2025 13:46 — 👍 8 🔁 3 💬 1 📌 0
So thrilled to see our pre-print online. This was an incredible team effort and I am so proud to have been part of this amazing study, special thanks goes to @nickywhiffin.bsky.social and @gregfindlay.bsky.social for their mentorship. Go check-out Nicky's thread hereunder:
11.04.2025 12:05 — 👍 6 🔁 3 💬 0 📌 0
In what is becoming a pretty well-oiled routine, Nicky reached out to see if we had Australian hits, and minutes later @cassimons.bsky.social had found a bunch of them. Now over 20 families diagnosed globally, many with unusual white matter changes - a distinct new recessive syndrome.
11.04.2025 11:29 — 👍 6 🔁 2 💬 1 📌 0
Not only is this seriously elegant science from @gregfindlay.bsky.social, @nickywhiffin.bsky.social and friends - using saturation editing to define variant impact in RNU4-2 - it also defines *another* new syndrome associated with this fascinating non-coding RNA gene.
11.04.2025 11:13 — 👍 27 🔁 7 💬 1 📌 0
high- and low-field (network) neuroimaging
Computational biologist using omics data (imaging, gene expression) to investigate the impacts of environmental contaminants on biological systems
Group Leader @EMBL-EBI (www.ewaldlab.org)
A platform for life sciences. Publications, research protocols, news, events, jobs and more. Sign up at https://www.lifescience.net.
Professor at #EPFL - #RNAVelocity inventor - Laboratory of Brain Development and Biological Data Science. Single-cell and spatial biologist studying brain development and lipids. #ERCStg investigator
Vice President, Biology @10xgenomics. Formerly Technology Innovation Lab @NYGCtech at New York Genome Center @nygenome. Views my own
Probabilistic machine learning to address questions in evolution and health #EvolutionaryMedicine. PI at the Centre for Genomic Regulation, co-leading a group with Mafalda Dias. Previously Harvard.
Hola! We are the CRG, a research institute exploring the frontiers of biology in Barcelona. Som un centre CERCA. www.crg.eu
Computational biologist @HelmholtzMunich, prof @TU_Muenchen & associate PI @sangerinstitute. Dad of 4 and mountain lover. Department news, see @CompHealthMuc
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, Compute Acceleration in biotech // http://albertvilella.substack.com
Researcher @bayraktar_lab @teichlab @steglelab.bsky.social @sangerinstitute.bsky.social | Using models & AI to study cells, cell circuits & brains 🧠 | #SingleCell+spatial | 🌍+🇺🇦
2025 Schmidt Science Fellow
CDB UCL London and Gurdon Institute Cambridge
Wilson, Simons and Norden labs
Developmental, Systems and Synthetic Biology
Prev: Briscoe, Crick Institute; Zernicka-Goetz and Thomson, Caltech; Banerjee and Goehring, UCL
RNA Bioinformatics Postdoc at Crick Institute/ UK DRI @ King's College London w/ Jernej Ule (@ulelab) and Ben Blencowe | Co-founder of Londonomics Network | Co-creator of Flow.Bio
Professor at KTH, NY Genome Center, SciLifeLab, working on functional genomics and human genetics.
Neuro and Developmental biologist. PostDoc at day. Supervillain at night. He/him #BlackLivesMatter #TransRightsAreHumanRights
@mads100tist@mastodon.social
postdoc @ http://debaerelab.com @ https://fungenlab-ugent.be | 3D genome, gene regulation, structural variation | retina, neurodev, space!
Clinic-minded genome + epigenome editor. Professor of Molecular Therapeutics, UC Berkeley. Director for Technology and Translation, Innovative Genomics Institute, ibid.
Beatles fan.
Art work credit: Fyodor Vasiliev "A Meadow in the Rain" (1872).
Molecular geneticist | Neurogenetics | repeat expansions | chromosome X | snRNAs | and more
Executive Director EMBL. I have an insatiable love of biology. Consultant to ONT and Cantata (Dovetail)