
Wrapping up an amazing 4th annual Mass Spec Day @jhu.edu is @jyates.bsky.social with a talk on the mechanism of CF. “Every day is mass spec day!” Thank you to our speakers, sponsors, and my fellow co-organizers. Looking forward to seeing yall @asms.org in #Baltimore!
30.05.2025 20:32 — 👍 2 🔁 0 💬 0 📌 0
Wouldn’t have been possible without the indefatigable @leunglab.bsky.social a brilliant co-organizer! Very pleased the day was such a success. Special
Thanks to student speakers who really took things up a notch!
11.05.2025 19:49 — 👍 2 🔁 0 💬 0 📌 0
Though putting on FCBIS 2025 was a lot of work, I think it was worth it and hearing this from friends is so satisfying! I’m glad we could bring our community together for a day of exchange, learning, and good times! Next year in Newark DE!
11.05.2025 19:05 — 👍 1 🔁 0 💬 0 📌 0
Finally, I am happy to say that I am trying to relocate to @bsky.app after a science social media hiatus. Looking forward to reconnect with you here. Please follow us if you find our work on protein folding interesting! 9/9
27.03.2025 15:00 — 👍 0 🔁 0 💬 0 📌 0
We have benefitted immensely from collaboration with Ed O'Brien, and this project was spearheaded by the remarkable team of Yang Jiang and
@friedlab.bsky.social's April Xia. Thanks to NSF and NIH for their support of this work. 8/9
27.03.2025 15:00 — 👍 0 🔁 0 💬 1 📌 0
To summarize, we think that entangled states represent a class of misfolding that can explain complex refolding kinetics & why some proteins do not refold efficiently. Since they are not easily caught by chaperones, they may pose dangers to our cells too. 7/9
27.03.2025 15:00 — 👍 0 🔁 0 💬 1 📌 0
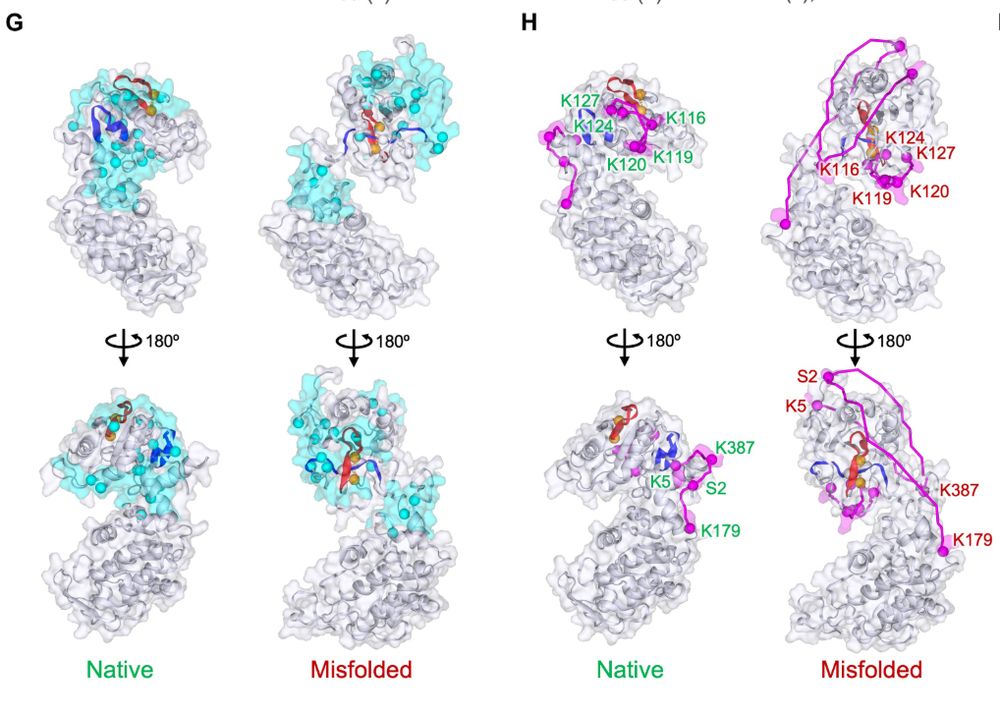
We also used two types of structural mass spectrometry, LiP-MS and XL-MS, to characterize the topologically misfolded form of PGK and showed that these features match the entangled conformations from simulations quite well. 6/9
27.03.2025 15:00 — 👍 0 🔁 0 💬 1 📌 0

We showed this in a few ways. First, Yang ran extensive simulations on PGK, showing it gets entangled, and if you take the trajectories away with entanglements the stretched exponential signature also disappears. 5/9
27.03.2025 15:00 — 👍 0 🔁 0 💬 1 📌 0

But if this is true, what are these various non-native states that cannot just sort themselves out and get back on the normal folding path? We think they are "entangled states," or conformations with non-covalent lasso entanglements. 4/9
27.03.2025 15:00 — 👍 0 🔁 0 💬 1 📌 0

Physically, this could correspond to a scenario of a rough free energy landscape with many non-native minima that do not easily interconvert and which have a diverse spectrum of rate constants. 3/9
27.03.2025 15:00 — 👍 1 🔁 0 💬 1 📌 0

Where do we begin? In the late 90s, protein folders noticed that not all proteins fold with expected single-exponential kinetics. The enzyme from glycolysis, PGK, appears to follow a stretched exponential function instead. 2/9
27.03.2025 15:00 — 👍 0 🔁 0 💬 1 📌 0
Hey #proteinfolding #biophysics #TeamMassSpec #proteomics people: After a long social media hiatus, @friedlab.bsky.social is back on @bsky.app. Looking forward to connecting with you all in this space, and filling my day with less alt-right/crypto/AI-nonsense!
21.03.2025 12:56 — 👍 1 🔁 0 💬 0 📌 0
America’s first research university. Leading discovery and sharing knowledge to better the world since 1876. With campuses & centers in Baltimore & around the world.
John Yates studies proteins using the coolest method on the planet, mass spectrometry. Thoughts and musings are my own. EIC of the Journal of Proteome Research. https://www.scripps.edu/yates/
All views and opinions expressed are my own.
Neurobiologist, Professor, fond of reading, cats, thrillers, and food
Computational physicist at https://peptone.io
PhD @GroupParrinello, PostDoc @franknoe.bsky.social
Disordered Proteins, AI for Science, Molecular Dynamics, Enhanced Sampling
🔗 https://scholar.google.com/citations?user=fnJktPAAAAAJ
Professor. Science and social about my lab at Johns Hopkins University working on PARP and RNA biology. Views are my own.
Assistant Professor of Physiology at the University of Pennsylvania | studying long-lived mitochondrial proteins, organelle stability, mtDNA fidelity in neurons and oocytes
www.bombawarczaklab.com
Alternating between running a lab and taking care of the kids. Tweets mostly science related.
Membraneless Organelles in Bacteria: Structure, Dynamics, Function, and Engineering. Assistant Professor @scripps.edu.
https://laskerlab.org/
Assistant Professor at UNC Biochemistry and Biophysics. Unraveling mysteries of disordered and dynamic proteins with NMR and biophysics. Views are my own.
www.berlowlab.org
Assistant professor of Chemical Engineering at USC in Los Angeles. My research group focuses on protein-lipid biophysics and biological membrane engineering.
Assistant Professor, Princeton University | biomolecular condensates + computer simulations | https://josephgroup.princeton.edu (she/her)
Professor at Chalmers in Sweden. Biophysical research on proteins, with focus on amyloids and metals. Fights for DEI. Crazy in good ways😀
🏳️🌈 Assoc. Prof @Max Perutz Labs Vienna, Biochemist & Structural Biologist, #RNA & #protein, Tennis addict, 🐈❤️,all time skeptic 🤨
Student-run THG Lab account @UC Berkeley. We develop physics-based and machine learning-based models for various systems.
Protein and coffee lover, father of two, professor of biophysics and sudo scientist at the Linderstrøm-Lang Centre for Protein Science, University of Copenhagen 🇩🇰
Promoting the study of the physical principles and mechanisms behind how living organisms survive, adapt, and grow.
Join now: https://go.aps.org/4eCIyvD
Reporter/editor/narrative coach @Bloomberg.com. Co-author, Unbelievable. Law school dropout/honorary doctor. Formers: ProPublica, Marshall Project, Seattle Times. Perturbable.
https://kenarmstrongwriter.com/
Husband, Father, Grandfather, Datahound, Dog lover, Fan of Celtic music, Former NIGMS director, Former EiC of Science, Stand Up for Science advisor, Shenanigator, Pittsburgh, PA
NIH Dashboard: https://jeremymberg.github.io/jeremyberg.github.io/index.html
Professor @BostonCollege
Chemical Biology, Synthetic Biology, Directed Evolution, Expanding the genetic code https://sites.google.com/a/bc.edu/thechatterjeelab/home
https://scholar.google.com/citations?user=zeSNxNcAAAAJ&hl=en







