Copious amounts of conditioner required. Costco amounts.
02.02.2026 23:16 — 👍 0 🔁 0 💬 0 📌 0Jess Heebner
@jessheebner.bsky.social
Deep Learning CryoET Image Analysis Specialist, cat lover, dog lover, mother to a 5yo tyrant
@jessheebner.bsky.social
Deep Learning CryoET Image Analysis Specialist, cat lover, dog lover, mother to a 5yo tyrant
Copious amounts of conditioner required. Costco amounts.
02.02.2026 23:16 — 👍 0 🔁 0 💬 0 📌 0
Good morning here is a gofundme to support three kids left behind when their mom was abducted. One of the kids has urgent medical needs. Please fill it up! www.gofundme.com/f/urgent-aid...
20.01.2026 16:29 — 👍 2 🔁 3 💬 0 📌 0I'm so happy to have gotten the opportunity to work on this project and I'm SO excited that they selected our image for the cover!
09.01.2026 19:20 — 👍 14 🔁 7 💬 0 📌 0JOB ALERT! 📢 For the scientists who love science communication: we’re hiring at @kurzgesagt.org !🌍 Come join our team, the vibes are impeccable.
✍️ Scriptwriter (Remote) kurzgesagt.org/careers/scri...
🔍 Fact-Checker & Researcher (Munich/Berlin/Remote) kurzgesagt.org/careers/seni...
When you get the reputation of being the guy with the encouraging words on New Year's Eve, it can start to come through as a little pressure -- what if the situation on the ground is worse than usual? what if people are more scared than they usually are, and with cause? what use are good vibes then?
31.12.2025 23:21 — 👍 1935 🔁 427 💬 18 📌 106
This might be my favorite piece of conference swag ever. My 5yo is obsessed with it. #cellbio2025 @jcb.org
13.12.2025 01:05 — 👍 14 🔁 2 💬 0 📌 1If you're passionate about #SciArt and #SciComm, don't miss our exciting session today at #ASCB #CellBio2025! Thank you @ascbiology.bsky.social!
07.12.2025 18:53 — 👍 10 🔁 3 💬 0 📌 0Do I need to go fill you a box from this hah
07.12.2025 16:53 — 👍 1 🔁 0 💬 1 📌 0What are your plans for #cellbio2025 ? Aside from eating too much at Reading Terminal Market and wandering the Christmas Market in my spare time... Come find me to chat all things segmentation (and come see my work displayed along the back wall of the Thermo Fisher booth!)
06.12.2025 18:31 — 👍 7 🔁 0 💬 0 📌 0Going to #ASCB2025 in Philly? Come find me! I'll be haunting the Thermo Fisher booth and wandering the conference. I'd love love love to talk all things segmentation! Let's get coffee. Show me your data. Reach out!
04.12.2025 01:01 — 👍 5 🔁 1 💬 0 📌 0This is pretty prevalent in how people communicate science in talks too. So many talks at conferences are just so stilted and dry and they don't have to be. You can communicate complicated topics accessibly to even other experts. But in my experience it's not taught well or at all in school.
18.11.2025 23:30 — 👍 1 🔁 0 💬 0 📌 0I'll be there all conference!
16.11.2025 15:08 — 👍 1 🔁 0 💬 0 📌 0📣 New paper alert! Just out in Cell Reports! pubmed.ncbi.nlm.nih.gov/40644298/
Thrilled to share that we have discovered a brand-new anti-phage defense system! Bacteria have evolved various defense strategies (CRISPR etc) to counter phage attacks. We found a new one - fascinating and dramatic
⚔️🦠❄️🔬
Sounds like you need a spreadsheet organization system too...
17.07.2025 00:46 — 👍 1 🔁 0 💬 0 📌 0NEW! Hydra Bio Plasma-FIB features AI protocols for bio specimens. Capture high-res 3D data from cryo-preserved or resin-embedded samples & generate high-quality cryo lamellae. Register for June 24 webinar to learn from users how they've enhanced their #volumeEM and #teamtomo prep. ter.li/d8b427
30.05.2025 16:47 — 👍 11 🔁 2 💬 1 📌 0Can't wait to try this out! I have loved seeing my segmentations come to life in Blender
23.05.2025 23:05 — 👍 5 🔁 0 💬 0 📌 0www.wehi.edu.au/careers/make...
WEHI is looking for new lab heads! New and experienced lab heads welcome. Check out the ad, consider applying and share. We’re a collaborative bunch, in Australia, with fabulous technology, great colleagues at a values-driven organisation. @wehi-research.bsky.social
Hey #TeamTomo,
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
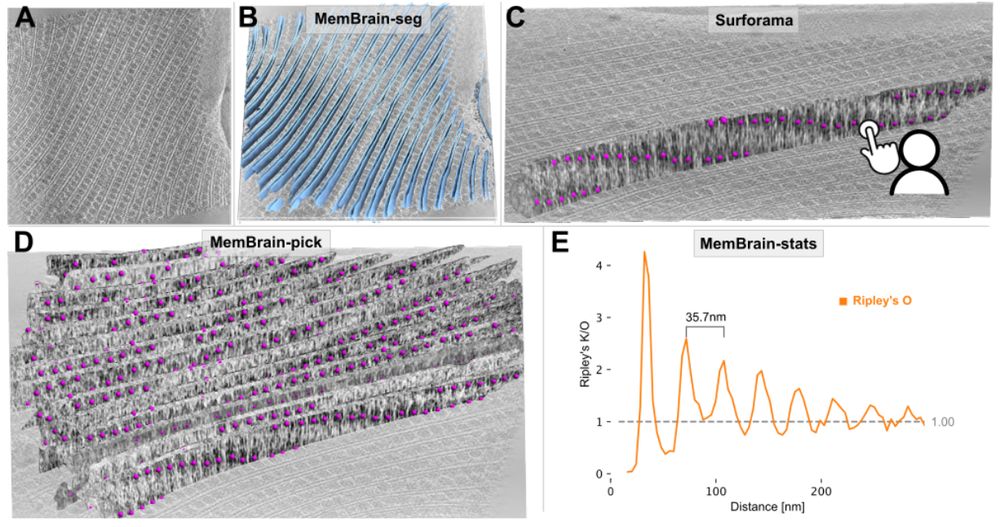
Figure 4. MemBrain v2 end-to-end workflow detects periodic phycobilisome organization. A: Raw tomogram slice of EMD-31244. B: Out-of-the-box MemBrain-seg segmentation (light blue). C: A single membrane instance can be visualized in Surforama and manually annotated with GT phycobilisome positions (magenta). D: MemBrain-pick localizes particles (trained with data from C) on all membranes in the tomogram. E: MemBrain-stats computes Ripley’s O statistic using the positions from D with a bin size of 5nm. The distance between peaks (35 nm) was measured to estimate chain unit spacings.
We have updated our #MemBrain v2 preprint with a lot more details about the MemBrain-pick and MemBrain-stats modules, as well as some application examples!
Stay tuned for the upcoming thread by lead author @lorenzlamm.bsky.social! 🧠🧵
#CryoET #TeamTomo
www.biorxiv.org/content/10.1...

So thrilled to unveil the Krios 5 Cryo-TEM for studying cellular structures and molecular interactions. Thank you to the many, many teammates that made this possible. Watch the video! Watch the more & sign up for virtual event on May 14: ter.li/ueeqfs
www.linkedin.com/posts/thermo...
We're on the cover of @science.org this week. Awesome work by @florentwaltz.bsky.social. #CryoET is reaching crazy resolutions inside cells, and this is just the beginning. Fantastic interpretation of the proton motive force by @verenaresch.bsky.social. Always a pleasure making #SciArt with you!🧪🧶🧬🌾
20.03.2025 22:37 — 👍 308 🔁 69 💬 16 📌 2What a gorgeous animation!
21.03.2025 01:16 — 👍 2 🔁 0 💬 0 📌 0When I was in grad school I would have loved to learn about roles for bioimage analysis outside of academia because I was struggling to figure out a career path for myself. So personally I'd find it interesting to include stuff like that in the panel!
14.03.2025 04:08 — 👍 1 🔁 0 💬 0 📌 0Things always get worse before they get better..something something pithy... I'm reposting this because the situation has become more urgent and exponentially more stressful. <3
06.03.2025 01:18 — 👍 2 🔁 0 💬 0 📌 0I'm in the process of segmenting all of the tomograms as well! We should look into the best way for sure!
28.02.2025 22:54 — 👍 1 🔁 0 💬 1 📌 0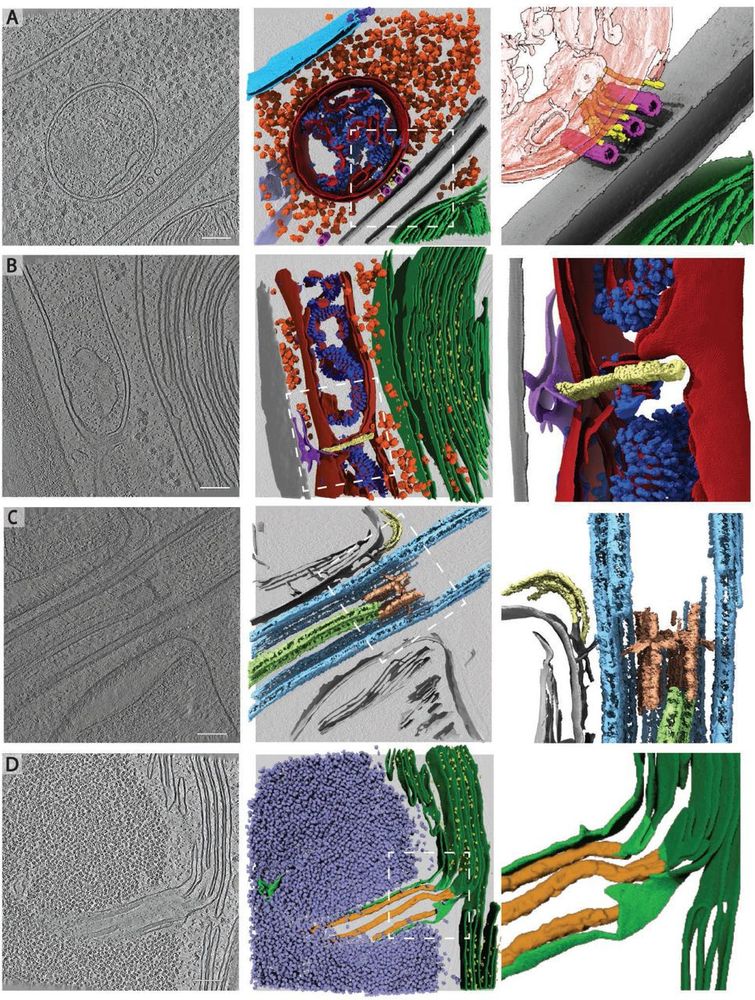
Rare cellular events observed in the cryo-ET dataset. Slices through tomograms (left) with corresponding 3D segmentation (middle and right). Right panels show enlarged views of the boxed regions in the middle panels. A) Mitochondrion at the plasma membrane, tethered to microtubules by thin filaments of unknown identity. Segmented classes: nuclear envelope (light blue), nuclear pore complex (dark blue), 80S ribosomes (orange), mitochondrial membranes (dark red), ATP synthase (royal blue), plasma membranes (grey), microtubules (pink), filaments (yellow), endoplasmic reticulum (purple). Zoomed-in panel shows thin filaments running between microtubules and the mitochondrion at the plasma membrane. Note that this tomogram contains two closely appressed cells, and thus, two plasma membranes with cell walls between. B) Mitochondrial fission event with ER membrane interactions. Segmented classes: mitochondrial membranes (dark red), ATP synthase (royal blue), 80S ribosomes (orange), thylakoid membranes (green), PSII (bright yellow), cell wall (grey), ER (purple), fission site containing filamentous structures perpendicular to the mitochondrial long axis (pale yellow). C) Ciliary transition zone between basal body and axoneme, including assembling IFT train and stellate structure77. Segmented classes: microtubule doublets (light blue), central microtubule pair (light green), stellate structure (pale red), IFT train (yellow). D) Pyrenoid tubule extending from the thylakoids into the phase-separated Rubisco matrix of the pyrenoid. Minitubules originate from thylakoid membranes82. Segmented classes: thylakoid membranes (dark green), PSII (yellow), pyrenoid tubule (lime green), Rubisco (lavender blue), minitubules (orange). Scale bars in A-D: 100 nm. Related to Fig. 2C-H.
To all #TeamTomo #CryoET and #Chlamydomonas aficionados: we have updated EMPIAR-11830 with a bug fix concerning some cryo-CARE denoised tomos as well as additional files and metadata! 🎉👩🏽💻
www.ebi.ac.uk/empiar/EMPIA...
A little thread about what's new... 🧵 1/n
Excited to see this out!!
05.02.2025 14:09 — 👍 6 🔁 0 💬 0 📌 0I typically stick to science/cryoem posts here, but this is very important to me as this is my brother and his family. I want to see them safe. <3
04.02.2025 01:31 — 👍 3 🔁 0 💬 0 📌 0Finally out! Working with this team was a fantastic learning experience. Also I was excited to determine the in situ clathrin structure for this project! That’s my first EM-structure upload to EMDB. Can I tell my mom I am a structural biologist now? 😁 #cryoET
06.01.2025 18:00 — 👍 38 🔁 4 💬 1 📌 1Thanks Alicia! I am also happy to have helped finalize this work. I hope computational biologists will find the repo useful. We need to share more annotations in accessible format, the algorithms are hungry 😄 #teamtomo, consider sharing your data via pull requests—let’s grow this together! 🤝
07.01.2025 13:18 — 👍 5 🔁 2 💬 0 📌 0