
Happy to present the final version of our Cadherin and gastruloid manuscript, online at @cp-cellreports.bsky.social.
www.cell.com/cell-reports...
Initially in BiorXiv (www.biorxiv.org/content/10.1...) 1/12
@shamipourshayan.bsky.social
Fascinated by how embryos get their shapes and patterns 🐠 SNSF Ambizione junior group leader @UZH Postdoc @Pelkmans lab | PhD @CPHeisenberg lab

Happy to present the final version of our Cadherin and gastruloid manuscript, online at @cp-cellreports.bsky.social.
www.cell.com/cell-reports...
Initially in BiorXiv (www.biorxiv.org/content/10.1...) 1/12
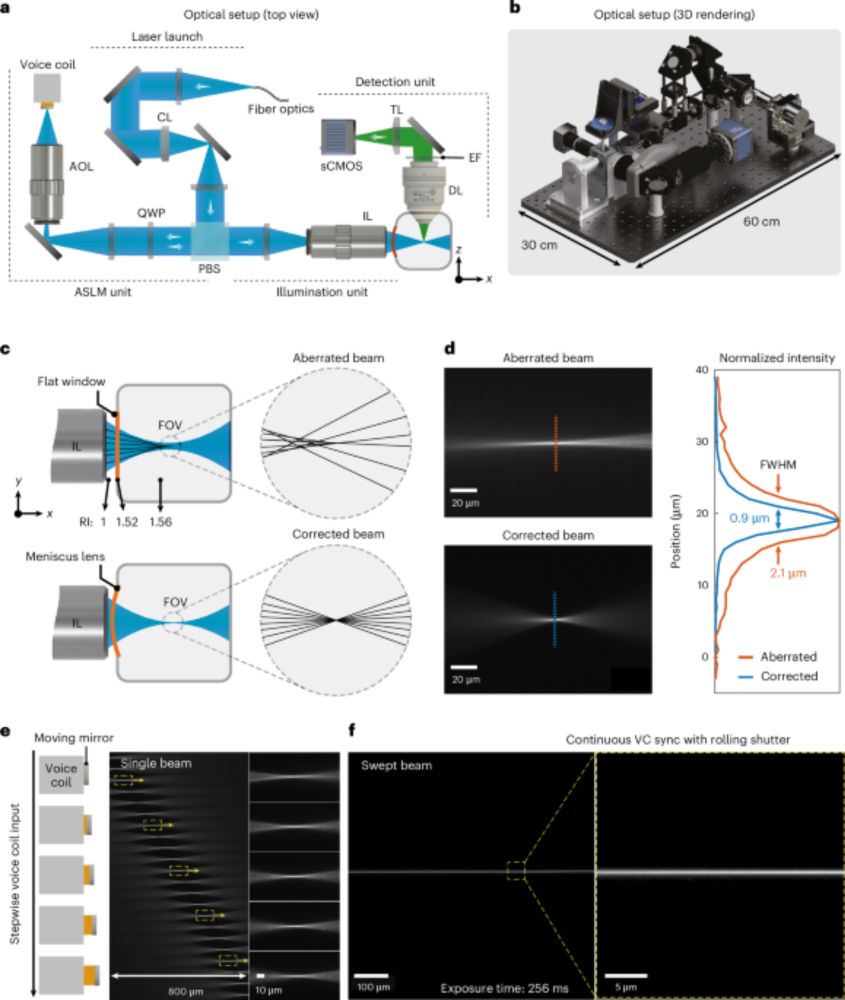
🎉 I am very glad to share that our paper on the rapid isotropic light-sheet microscope has just been published in Nature Biotechnology @natbiotech.nature.com.
🔗 Read the full article:
www.nature.com/articles/s41...

Why do whales and elephants have much lower rates of cancer than expected?
"Species with a lower cancer prevalence and mortality
risk are those with a higher presence of cooperative and caring habits, while the opposite is found....:"
www.science.org/doi/10.1126/...
#ZebrafishZunday: Cytoplasmic (or ooplasmic) streaming leads to the segregation of embryo from yolk granules. Credit to @shamipourshayan.bsky.social & @heisenbergcplab.bsky.social. 🧪
16.11.2025 09:31 — 👍 55 🔁 8 💬 1 📌 2@sushantbangru.bsky.social and @rdiegmiller.bsky.social new review on regeneration is out in Nature Reviews Molecular Cell Biology. A fun piece to write and another great collaboration with @kenposs.bsky.social.
13.11.2025 20:48 — 👍 24 🔁 12 💬 0 📌 0We spy the latest works from the labs of @marymullins.bsky.social, Margot Kossman Williams (x2), @bruceappel.bsky.social, Vladimir Korzh, @ssumanas.bsky.social, @campaslab.bsky.social, @kenposs.bsky.social, @emiliapsantos.bsky.social among others! Do check it out!
13.11.2025 13:33 — 👍 5 🔁 1 💬 0 📌 0
How can we organize current theoretical approaches for developmental biology - from information to dynamical systems & GRNs - into a common framework?
We propose to think along Marr's 3 levels: computational problem, algorithm, implementation
Check out our review:
arxiv.org/abs/2510.24536

Interested in polarity and mechanics during epithelial morphogenesis? Considering joining us! Students to be hired initially as Research Associates. Postdocs encouraged to obtain fellowship, and we help/train you to do that. Apply to tinyurl.com/y6x489t5. Deadline Dec 15.
Please repost!
I’ll be speaking about the mechanical development of crocodile and tortoise head scales next Wednesday at 4pm for the @tlmcambridge.bsky.social seminar series. Subscribe to the mailing list to hear more about my work, and that of @maltemederacke.bsky.social!
lists.cam.ac.uk/sympa/subscr...
thanks a lot! 🙏 looking forward to seeing you next week :)
29.10.2025 14:28 — 👍 1 🔁 0 💬 0 📌 0Special thanks to our collaborators
@nvastenhouw.bsky.social and Darren Gilmour labs for all the critical feedback, and to @snsf.ch and @embo.org for funding support that made this work possible 🥂
Altogether, 3D-4i enables systematic analysis of molecular and spatial heterogeneity across scales, providing a new framework for understanding how coordinated regulation emerges in complex multicellular systems. 🌱💡
29.10.2025 14:20 — 👍 4 🔁 0 💬 1 📌 0
With cell cycle phase and other ZGA-associated factors measured in our dataset, we show that this heterogeneity does not arise from stochastic fluctuations and that we can predict the transcriptional activity of individual cells with high accuracy, linking cellular state and transcriptional output.
29.10.2025 14:20 — 👍 2 🔁 0 💬 1 📌 0
Since cell cycle plays a key role in transcriptional activity, we developed a computational pipeline to infer cell cycle phase directly from our multiplexed imaging datasets of fixed embryos. 🤩
29.10.2025 14:20 — 👍 2 🔁 0 💬 1 📌 0Using 3D-4i, we can visualize multiple protein and protein-state markers causally linked to regulating this process, both at single-cell level and across all embryos.
29.10.2025 14:20 — 👍 2 🔁 0 💬 1 📌 0
We applied 3D-4i to study the process of zygotic genome activation (ZGA), the stage where the embryo’s own genome takes control of development, which is highly heterogeneous at the single-cell level. 🤔
29.10.2025 14:20 — 👍 2 🔁 0 💬 1 📌 0Very happy to share our latest work extending iterative immunofluorescence to in toto imaging of early zebrafish embryos (3D-4i), integrated with a 3D-dedicated image analysis pipeline! 🐟🔬📊
Huge kudos to Max Hess for this PhD milestone! 💪
@lucaspelkmans.bsky.social shorturl.at/cO0u7
Let's zoom in!

With cell cycle phase and other ZGA-associated factors measured in our dataset, we show that this heterogeneity does not arise from stochastic fluctuations and that we can predict the transcriptional activity of individual cells with high accuracy, linking cellular state and transcriptional output.
29.10.2025 10:52 — 👍 0 🔁 0 💬 0 📌 0
Since cell cycle plays a key role in transcriptional activity, we developed a computational pipeline to infer cell cycle phase directly from our multiplexed imaging datasets of fixed embryos. 🤩
29.10.2025 10:52 — 👍 0 🔁 0 💬 1 📌 0Using 3D-4i, we can visualize multiple protein and protein-state markers causally linked to regulating this process, both at single-cell level and across all embryos.
29.10.2025 10:52 — 👍 1 🔁 0 💬 1 📌 0
We applied 3D-4i to study the process of zygotic genome activation (ZGA), the stage where the embryo’s own genome takes control of development, which is highly heterogeneous at the single-cell level. 🤔
29.10.2025 10:52 — 👍 0 🔁 0 💬 1 📌 0
With cell cycle phase and other ZGA-associated factors measured in our dataset, we show that this heterogeneity does not arise from stochastic fluctuations & that we can predict the transcriptional activity of individual cells with high accuracy, linking cellular state and transcriptional output.
29.10.2025 10:26 — 👍 0 🔁 0 💬 0 📌 0
Since the cell cycle plays a major role in transcriptional activity, Max developed a computational pipeline to infer cell cycle phase directly from our multiplexed imaging datasets of fixed embryos 🤩
29.10.2025 10:26 — 👍 0 🔁 0 💬 1 📌 0Vertebral skeletal diversity in mammals is remarkable. How do the differences between vertebral size and shape develop and evolve? See how we tackled this question in our paper published in @natcomms.nature.com today!
10.10.2025 18:46 — 👍 49 🔁 15 💬 2 📌 1Today my @nytimes.com colleagues and I are launching a new series called Lost Science. We interview US scientists who can no longer discover something new about our world, thanks to this year‘s cuts. Here is my first interview with a scientist who studied bees and fires. Gift link: nyti.ms/3IWXbiE
08.10.2025 23:29 — 👍 4738 🔁 1837 💬 142 📌 83New preprint from the lab! We discovered a transient fluidization in the basal region of human forebrains by tracking microdroplets in cerebral organoids.This “basal fluidization”, absent in gorilla and mouse, may contribute to greater surface expansion in human forebrains
1/
doi.org/10.1101/2025...


🧠 Join us @uzh-ch.bsky.social ! We are a new lab looking for PhDs, Postdocs & Master’s students interested in studying how human neural systems emerge & go awry using brain organoids, light-sheet imaging & genomics. 🚀 Deadline for LSZGS 1st Nov! www.lifescience-graduateschool.uzh.ch/en.html.
07.10.2025 11:57 — 👍 24 🔁 12 💬 0 📌 0
First, check out my recent bioRxiv preprint w/ @oweinerlab.bsky.social: www.biorxiv.org/content/10.1...
We find contractility shifts the proportion of implantation-competent embryos from young and aged females. Keep reading this for more info!
Our latest preprint! Into morphogenesis, Yap, ECM, signaling, vertex models, feedbacks/robustness? There is something here for everyone. We discovered a positive feedback loop that extends inner ear canals, and a built-in mechanism that shuts it down when morphogenesis is done. tinyurl.com/464wsjhd
26.09.2025 12:55 — 👍 77 🔁 27 💬 4 📌 2🚨 New paper out in @pnas.org! 🚨
We show that self-generated gradients allow heterogeneous cell mixtures to co-migrate efficiently over long distances —while optimizing their physical interactions!
(See below for 🧵)
www.pnas.org/doi/10.1073/... 🧪