I was also going to point out it being a bit slow, but was waiting to see my job complete first! Thanks for figuring it out. I also noticed that the tutorial config.yaml may have the wrong names for fields e.g. "tomo1" needs to be "tomo1_files" for it to run
27.10.2025 10:53 — 👍 1 🔁 0 💬 1 📌 0
Thanks!
16.10.2025 06:29 — 👍 0 🔁 0 💬 0 📌 0
Thanks!
15.10.2025 20:08 — 👍 0 🔁 0 💬 0 📌 0
Electron Microscopy Data Bank
Thanks! According to their stats page (www.ebi.ac.uk/emdb/statist...) I should be at around 37% right now, but we'll have to see how the rest of the year goes! 📈
15.10.2025 17:56 — 👍 3 🔁 0 💬 0 📌 0
Thanks!
15.10.2025 14:32 — 👍 0 🔁 0 💬 0 📌 0
Thanks! I didn't see h44 repositioning despite directly looking for it. Could be the P-tRNA, but I also suspect IF3 might be involved in clamping it down in some way (>90% of 30S had IF3).
15.10.2025 14:31 — 👍 2 🔁 0 💬 0 📌 0
Thanks! Although my wrists might disagree :)
15.10.2025 14:15 — 👍 1 🔁 0 💬 0 📌 0
Sadly not :( I got most of the model IDs sequential, but the maps were a different story
15.10.2025 11:59 — 👍 2 🔁 0 💬 0 📌 0
Thanks!
15.10.2025 11:40 — 👍 1 🔁 0 💬 0 📌 0
Thanks!! Very true, and things are getting more and more doable- I think it'll be quite a different place in a few years.
15.10.2025 07:03 — 👍 0 🔁 0 💬 0 📌 0

Figure 1 from the preprint
Check out our preprint! With new molecular mechanisms, 140 subtomogram averages, and ~600 annotated cells under different conditions, we @embl.org were able to describe bacterial populations with in-cell #cryoET. And there’s a surprise at the end 🕵️
www.biorxiv.org/content/10.1...
#teamtomo
15.10.2025 06:26 — 👍 113 🔁 35 💬 8 📌 4

The small GTPase Ran defines nuclear pore complex asymmetry
Nuclear pore complexes (NPCs) bridge across the nuclear envelope and mediate nucleocytoplasmic exchange. They consist of hundreds of nucleoporin build…
My PhD paper is in press @cellpress.bsky.social and now online: www.sciencedirect.com/science/arti...!
Contents: weird #NPCs in yeast and flies, #CLEM, in situ #cryoET and more!
Made possible by the amazing @becklab.bsky.social team at @mpibp.bsky.social and collaborators! #TeamTomo
18.08.2025 16:26 — 👍 122 🔁 28 💬 6 📌 3
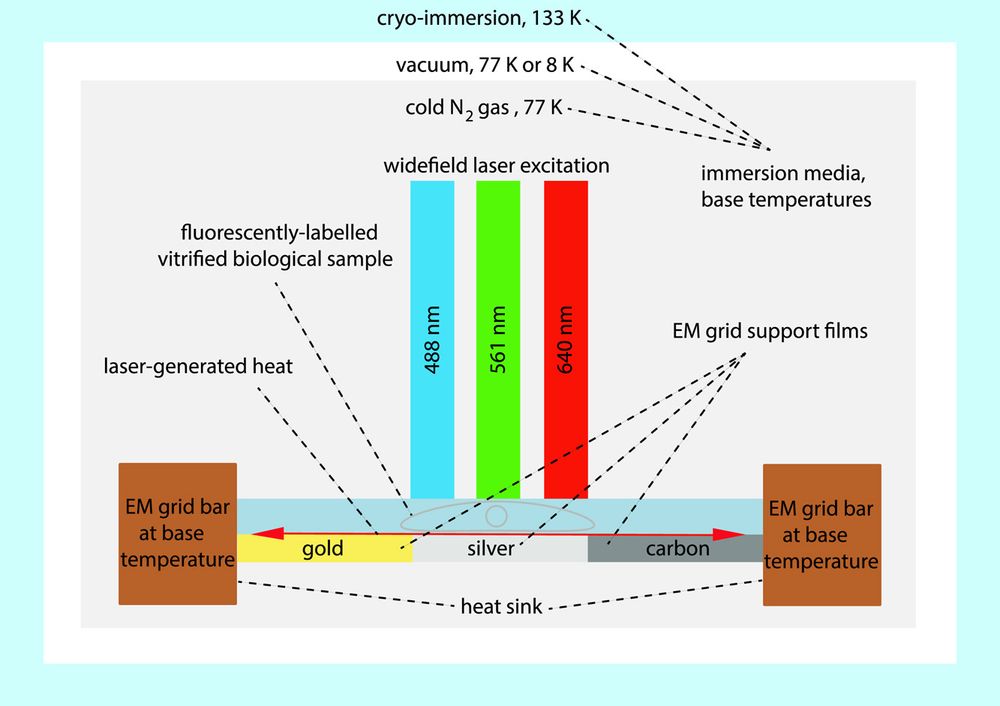
New paper from my colleague Soheil in the Mahamid and @jonasries.bsky.social groups looking at factors influencing laser devitrification on frozen #cryoem samples. With a far-red fluorophore, all-gold support grids will be especially good for correlative cryo-SMLM and EM!
doi.org/10.1016/j.js...
19.07.2025 02:32 — 👍 12 🔁 3 💬 0 📌 0
Are you into cryo-ET, in-cell discovery, protein translocation and folding or just cool integration of structural biology, proteomics and bioinformatics? Then see our new work:
www.biorxiv.org/content/10.1...
Thank you to all coauthors for your amazing work!
28.04.2025 04:43 — 👍 19 🔁 4 💬 2 📌 1
Associate prof @ Scripps Research
grotjahnlab.org
#teamtomo #mitochondriac
Structural neuroscientist || Associate Professor || UKRI Future Leader Fellow using #cryoEM #teamtomo to explore inside mammalian brains
@AstburyCentre @UniversityLeeds
EMDB Team Leader - EMBL-EBI. wwPDB Principal Investigator. Fathering, mixology & DIY enthusiast. #CryoEM #CryoET #firstgen. All opinions are my own.
Dean of Science, Professor of Chemistry, and Professor of Physiology & Cellular Biophysics at Columbia University | Single-Molecule Biophysics, Structural Biology, and Biochemistry | Diversity, Equity, and Inclusion in STEM
We are interested in life sciences, structural biology, translation, ribosomes, neurobiology and RNA therapeutics.
Location: RNA Therapeutics Institute, UMass Chan Medical School, Worcester, MA, USA
https://www.umassmed.edu/korostelev-lab/
#cryoem #teamtomo #volumeEM #thermofisherscientific
Yosemite is my happy place
enthusiast of transposable elements, genetic conflicts, small RNAs, Drosophila, and funky germline biology
running a lab at IMBA, Vienna BioCenter
https://www.oeaw.ac.at/imba/research/julius-brennecke
mRNA & cryo-EM enthusiasts at IMP Vienna. Posts are by lab members.
PhD student in Martin Beck’s group (MPI Biophysics)
Structural and mechanistic study of proteins, especially redox- and metalloproteins
We are the Savitski lab located @ EMBL using and developing proteomics methods for assessing the state of the proteome
Evolutionary cell biology @EMBL
evonuclab.org
keen on reverse transcription | also keen on spliceosomes | cryoEM dabbler
Assistant Member @MSKCC
Formerly post-doc @MIT
Formerly-formerly PhD @MRC_LMB
Formerly formerly-formerly Otago Uni
Kiwi 🥝 🇳🇿
https://wilkinsonlab.bio/
PhD Candidate at UBC Biochemistry with John Burke and Calvin Yip | CIHR Doctoral Scholar | Structural Biology | Cryo-EM | HDX-MS | Kinases and Phospholipids | 🇨🇦 | Seeking a postdoctoral position
CryoEM team @Genentech. Facility management, grid prep, screening, data collection, image processing, modelling, training. Opinions are my own.
CryoEM at Scripps Research
Postdoc in the Carter Lab at the MRC-LMB. EMBO fellow. Alumnus of the Brouhard Lab.
Neural Circuits and Behaviour in Drosophila @ MRC LMB. PI @ flyconnectome @ CamZoology and @ virtualflybrain. Cambridge, UK. Previously @gsxej.



