Genetic evaluation of CRISPR-Cas9 off-target effects from deleterious mutations on Drosophila male single X chromosome
#Drosophila
PubMed link
Genetic evaluation of CRISPR-Cas9 off-target effects from deleterious mutations on Drosophila male single X chromosome
#Drosophila
10.12.2025 07:23 — 👍 2 🔁 1 💬 0 📌 0
The story of how multiple RBPs cooperate to repress translation is now out in @cp-cellreports.bsky.social by @marcopayr.bsky.social and in a great collaboration with @hennig-lab.bsky.social!
www.cell.com/cell-reports...
07.11.2025 15:34 — 👍 40 🔁 14 💬 3 📌 0
Transcript-wide m6A methylation defines the efficiency of the cap-independent translation initiation https://www.biorxiv.org/content/10.1101/2025.11.25.690160v1
26.11.2025 02:17 — 👍 1 🔁 1 💬 0 📌 0
Analysis of Transcript Expression and Core Promoter DNA Sequences of Brain, Adipose Tissues and Testis in Human and Fruit Fly
#Drosophila
PubMed link
Analysis of Transcript Expression and Core Promoter DNA Sequences of Brain, Adipose Tissues and Testis in Human and Fruit Fly
#Drosophila
27.11.2025 12:23 — 👍 1 🔁 1 💬 0 📌 0
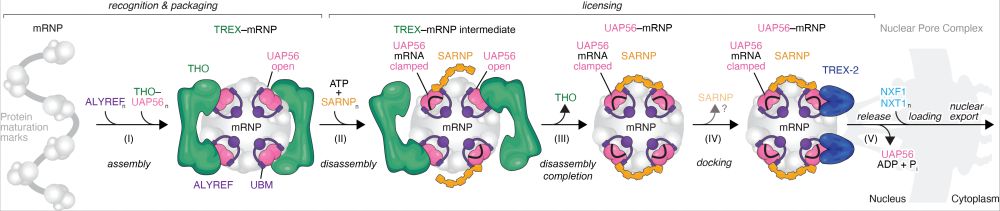
Finally out in @nature.com! We uncovered a mechanistic framework for a general and conserved mRNA nuclear export pathway. www.nature.com/articles/s41.... 1/
19.11.2025 23:21 — 👍 103 🔁 42 💬 3 📌 1

Functions of RNA m6A methylation at the molecular, genomic and organismal level: go.nature.com/484M6pB
Free to read here: rdcu.be/eR4zx
27.11.2025 12:51 — 👍 10 🔁 5 💬 0 📌 1

m⁵C-Rol-LAMP, a simple qPCR-based assay, quantifies m⁵C RNA methylation at single nucleotides and uncovers stress-regulated rRNA modification in bacteria. #LMU_muenchen #lifescimunich #RNA #CRC1309 bit.ly/43AWnr4
20.11.2025 13:54 — 👍 2 🔁 2 💬 1 📌 0

The missing heritability question is now (mostly) answered
Not with a bang but with a whimper
I wrote a little bit about the "missing heritability" question and several recent studies that have brought it to a close. A short 🧵
21.11.2025 22:33 — 👍 351 🔁 170 💬 14 📌 21

Ancient fossil reveals how plants and fungi first developed on land | Natural History Museum
A new fossil fungus discovered in Scotland shows evidence of plants and fungi sharing nutrients to survive on land.
A new fossil fungus discovered in Scotland shows evidence of plants and fungi sharing nutrients to survive on land.
The fossil, more than 400 million years old, offers hints about the origin of one of the greatest partnerships in the history of life on Earth.
www.nhm.ac.uk/discover/new...
20.11.2025 15:35 — 👍 105 🔁 49 💬 2 📌 1
Looking for a PhD in RNA biology? The deadline is approaching fast! This is a great opportunity to do a PhD together with an industry partner fully funded by @nrpdtp.bsky.social BBSRC Case studentship.
20.11.2025 08:05 — 👍 8 🔁 8 💬 0 📌 0
Single-nucleotide resolution mapping of m6A of zebrafish mRNAs in early embryonic development links m6A modifications to the maternal-to-zygotic transition https://www.biorxiv.org/content/10.1101/2025.11.19.688951v1
19.11.2025 16:33 — 👍 2 🔁 2 💬 0 📌 0
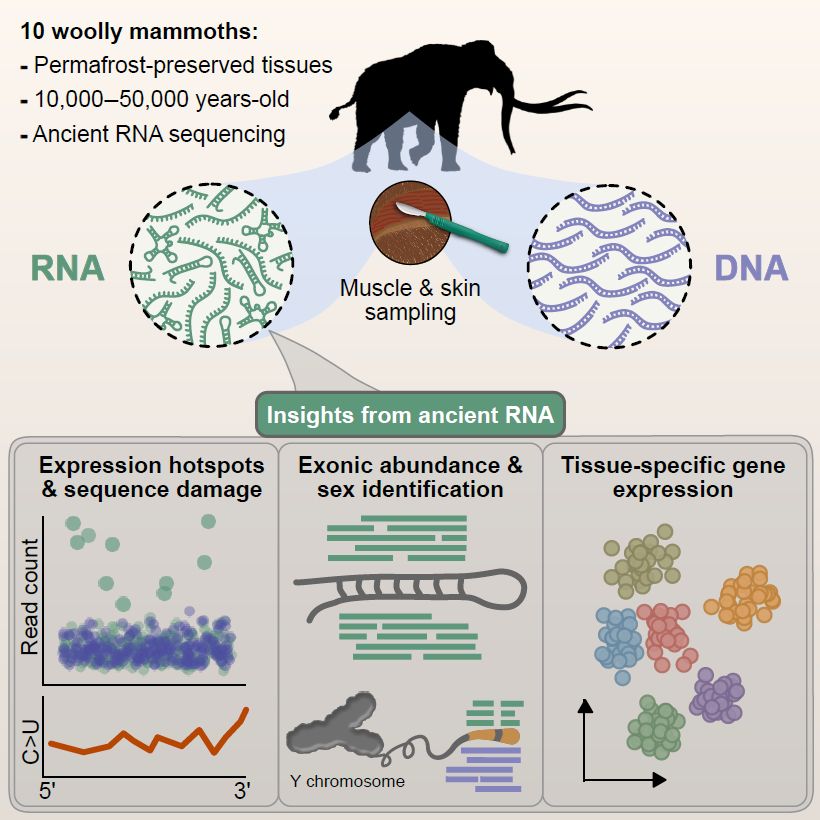
🦣🧬🦣🤯💥We are pleased to share our new paper about ancient RNA expression profiles from the Woolly Mammoth, now published in Cell @cellpress.bsky.social
www.cell.com/cell/fulltex...
If you want to know more, read the 🧵 below:
14.11.2025 16:08 — 👍 109 🔁 41 💬 1 📌 6
Control of gene output by intron RNA structure https://www.biorxiv.org/content/10.1101/2025.11.08.687378v1
10.11.2025 03:46 — 👍 6 🔁 2 💬 0 📌 2
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, AI/ML in biotech // http://albertvilella.substack.com
Molecular and computational biologist at Rutgers NJMS. Ramapo and UPenn alum.
Lecturer/ Assistant Professor in Advanced Cell Biology, University of Southampton, UK. Alumni: Stanford, Cambridge, Kings College London. 🇬🇧 🇮🇳 தமிழ் #FirstGen #NewPI. RNA Splicing Investigator at www.spliclab.co.uk
Flower Designer at Neoplants 🌺
Researcher at Binomica Labs
Degreeless Heathen
Petunia and Snapdragon Breeder
Open Lab Notebook: http://tinyurl.com/ATGCFFE
Statistical geneticist. Associate Prof at Dana-Farber / Harvard Medical School.
www.gusevlab.org
Professional scientist. I like reading books about history. Editor of the Bluesky edition of Gibbon's Decline and Fall of the Roman Empire.
http://historybooksreview.co.uk/
https://beautyscientist.github.io/GSDIANOZAS/
Studying why and how behavior evolves, from mosquitoes to mole-rats | Postdoc/Leon Levy Scholar @Columbia working with Ishmail Abdus-Saboor | PhD @Princeton with Lindy McBride | 麻布/東大 alum 🇯🇵 | yukihaba.github.io
PhD student at the Laboratory of #Chromatin Biology and #Epigenetics @IISERPune. Uncovering life’s secrets, one experiment at a time. #Zebrafish #GeneRegulation #Genomics #Bioinformatics #Devbio
https://orcid.org/0000-0003-3337-8280
Incoming postdoc in Claudia Bank’s lab. Promoters, emergence, fitness landscapes, dogs 🐶 and triathlon 🏊 🚲 🏃. He/him 🏳🌈.
timothyfuqua.com
Drosophila evolutionary and reproductive geneticist at Holy Cross. Seattle sports fan. MD spouse, preschool dad, SCOTUS junkie.
Retired researcher in Developmental Genetics, Evo-Devo. Drosophila. Polycomb Group. (PRC genes. Finder of "polyhomeotic"), Nuclear transplantation in the egg. Cell mosaics and chimeras. Epigenesis in Drosophila. CSIC --> CNRS. Hobby: mosaics.
Assistant Prof. @universitedeliege.bsky.social @frsFNRS @erc.europa.eu | Postdoc @rockefelleruniv.bsky.social | PhD @igbmc.bsky.social @unistra.fr | Studying RNA-protein machines ⚙️🏗 using cryo-EM ❄🔬
Automatically posts newly uploaded #Drosophila papers from Pubmed every hour.
Admin: @tkmkmym.bsky.social
Structural biologist and biochemist. CNRS researcher at CBM Orléans @cbm-upr4301.bsky.social. Interested in protein modifications & interactions. Also husband, dad of 2, friend, ☧. Personal website: msuskiewicz.github.io
Padre, curieux, scientist; sourdough engineer; evolutionary cell and systems biology; some attempts at synthetic biology and biomedical research;
Virologist, bioinformatician, NGS, photographer.
RNA is my special interest 🧬
Proteomics is growing on me 😬
🇳🇿🇬🇧British Kiwi based in Liverpool 📍
Post doc in the Emmott Lab👩🏻🔬👩🏻💻
🏳️🌈 She/They.
🧠 Neurodivergent ✨
rebee.co.uk
Plant small RNA and epigenomes enthusiast; recently repotted from Arizona to University of Oxford; academic mom






















