A complete map of human cytosolic degrons and their relevance for disease
Degrons are short protein segments that target proteins for degradation via the ubiquitin-proteasome system and thus ensure timely removal of signaling proteins and clearance of misfolded proteins fro...
In collaboration with the @lindorfflarsen.bsky.social group we release our map of degrons in >5,000 human cytosolic proteins with >99% coverage. A machine learning model trained on the data identifies missense variants forming degrons in exposed & disordered regions. Work led by @vvouts.bsky.social.
15.05.2025 11:56 — 👍 18 🔁 8 💬 0 📌 1
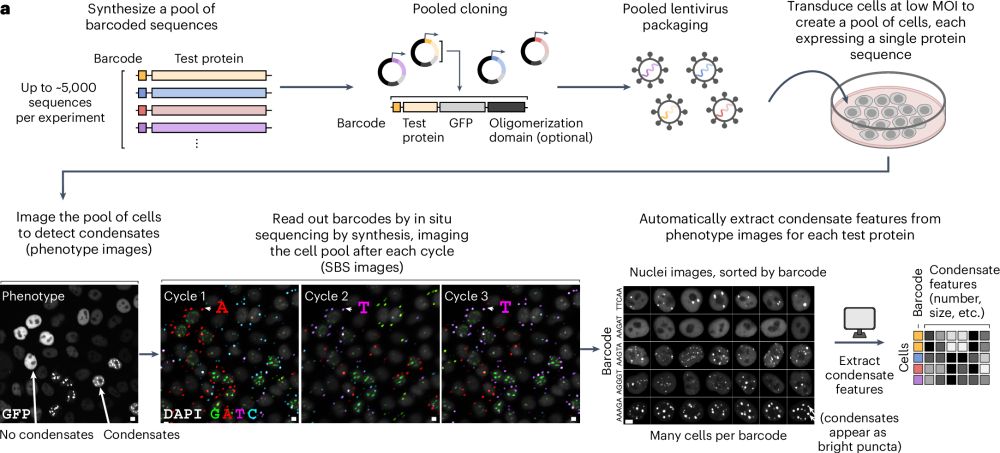
We are finally ready to share the preprint version about our use of mRNA-display to study what makes disordered proteins form condensates, at the proteome-scale!
Check it out here:
21.12.2024 16:10 — 👍 24 🔁 5 💬 1 📌 1
I am not quite sure that I can follow your question. We are indeed surprised that partitioning to DDX4N1 condensates appear to generalise to the behaviour of unrelated IDRs, so it most be sufficiently "promiscuous". Here, interactions seem to be better explained by partitioning rather than binding.
22.12.2024 07:26 — 👍 0 🔁 0 💬 0 📌 0
Thermodynamic Stability Modulates Chaperone-Mediated Disaggregation of α-Synuclein Fibrils https://www.biorxiv.org/content/10.1101/2024.12.19.629136v1
21.12.2024 13:48 — 👍 2 🔁 1 💬 0 📌 0
Check out @rasmusnorrild.bsky.social's work with Alex Buell and Joe Rogers developing and using Condensate Partitioning by mRNA-Display to probe phase separation of ~100.000 sequences, and @sobuelow.bsky.social's simulations to support and analyse the experiments
www.biorxiv.org/content/10.1...
21.12.2024 16:32 — 👍 36 🔁 8 💬 1 📌 0
We are finally ready to share the preprint version about our use of mRNA-display to study what makes disordered proteins form condensates, at the proteome-scale!
Check it out here:
21.12.2024 16:10 — 👍 24 🔁 5 💬 1 📌 1
Proteome-scale quantification of the interactions driving condensate formation of intrinsically disordered proteins https://www.biorxiv.org/content/10.1101/2024.12.21.629870v1
21.12.2024 15:50 — 👍 5 🔁 3 💬 0 📌 1
We also had the discussion a while back, and also ended up with the "signal above baseline" from laser scattering. Something like signal beyond 10 standard deviations of noise. But it does not help that you get "nano-cluster", or what else you call it, before full phase separation.
05.12.2024 12:17 — 👍 2 🔁 0 💬 1 📌 0
Protein Folder, father of two, married to @mariacaffarel.bsky.social
at UPV-EHU and DIPC
https://sites.google.com/view/biokt
stanford phd • nsf grf • jhu bs • deep learning for protein design & dynamics • oly weightlifter 🏋🏽♀️• 🇪🇬
gelnesr.github.io
Avid rower who sometimes thinks about biomolecular dynamics. Asst Prof @uwbiochem.bsky.social
https://waymentsteelelab.org/
Phase separation and disordered proteins!
Assistant Professor at UCLA
HHMI Hanna Gray Faculty Fellow
PhD: Stanford Biophysics; Postdoc: Broad Institute
Excited about RNA+proteins+IDPs, high-throughput experiments+computation
Designing proteins for better cancer immunotherapy | MD/PhD student @ WashU in the Nathan Singh Lab | UofSC Alum | #FlyEaglesFly
PhD student at MPINAT. Microtubules and coarse grained simulations.
Protein scientist in Copenhagen
Franco Lab @UCLA Exploring how to program dynamic behaviors in biological self-assembly 💡✨ We’re harnessing DNA and RNA to build smart, adaptive materials 🔬🧬 #Biomaterials #DNA #RNA
EPIGENETIC HULK SMASH PUNY GENOME. MAKE GENOME GO. LOCATION: NOT CENTROMERE, THAT FOR SURE
🇨🇦🇭🇰 @EPFL_en protein production + structure facility 🇨🇭| ex @ubc @uwaterloo @unige_en | here for #proteins #structuralbiology #xray #cryoem #plants #mountains #politics #transit #urbanism 🐱🐈 staying for the #hottakes | My views here🧪🧬🌱🔬🏳️🌈
Assistant Professor at U Arizona. Former: Postdoc with Gene Yeo at UCSD, Ph.D with Sua Myong at Johns Hopkins, BA/MA at Boston University. Cat lover, houseplant enthusiast, competitive gamer, aspiring foodie 🌈
Incoming assistant professor of chemistry at the Technical University of Denmark (DTU). Also at Jura Bio. machine learning, statistics, chemistry, biophysics
https://eweinstein.github.io/
Probabilistic machine learning to address questions in evolution and health #EvolutionaryMedicine. PI at the Centre for Genomic Regulation, co-leading a group with Mafalda Dias. Previously Harvard.
PhD in SynBio at MPI-Marburg, Researcher at prof. H. Niederholtmeyer group, TUM-CS
| Microfluidics | Cell-free systems | Biomolecular Condensates
🇱🇹 - 🇩🇪
Postdoc at ETH Zurich in the Prof. Paolo Arosio Lab at D-CHAB. Interested in protein aggregation and phase separation.
Explore the remarkable diversity of birds with Birds of the World, the world's leading collection of avian life histories published by the Cornell Lab of Ornithology in collaboration with a global editorial team of avian experts. birdsoftheworld.org.
Laboratory of Prof. Lewis E. Kay at the University of Toronto | Nuclear Magnetic Resonance of biomolecules | Est. 1992