🚨New preprint! We built a cell-free genomics platform (GATO-seq) to probe transcriptional regulation and discovered a “super pause” sequence that triggers a new Pol II active-site conformation. Huge shout-out to @robertovn.bsky.social for pulling off this monster of a project. tinyurl.com/superpause
19.02.2026 14:24 —
👍 49
🔁 12
💬 0
📌 1

🚨Preprint alert🚨 How does chromatin “architecture” form at CTCF sites? Our new preprint with @voslab.org and @andersshansen.bsky.social shows CTCF dimerization promotes nucleosome oligomerization on chromatin. tinyurl.com/CTCF-nucleos...
09.02.2026 15:04 —
👍 68
🔁 18
💬 0
📌 3
Happy to share part of my postdoctoral work at the @lucas.farnunglab.com lab. Great collaboration with @voslab.org and @andersshansen.bsky.social. “Structural basis for CTCF-mediated chromatin organization” www.biorxiv.org/content/10.6...
09.02.2026 15:22 —
👍 30
🔁 10
💬 0
📌 0
🧪🧬New preprint We present cryo-EM structures of reconstituted CTCF–nucleosome complexes, showing CTCF dimerization drives nucleosome oligomerization into defined higher-order assemblies. Disrupting CTCF–CTCF interfaces in mESCs reduces looping and impairs differentiation. tinyurl.com/CTCF-nucleos...
09.02.2026 12:54 —
👍 123
🔁 52
💬 4
📌 3
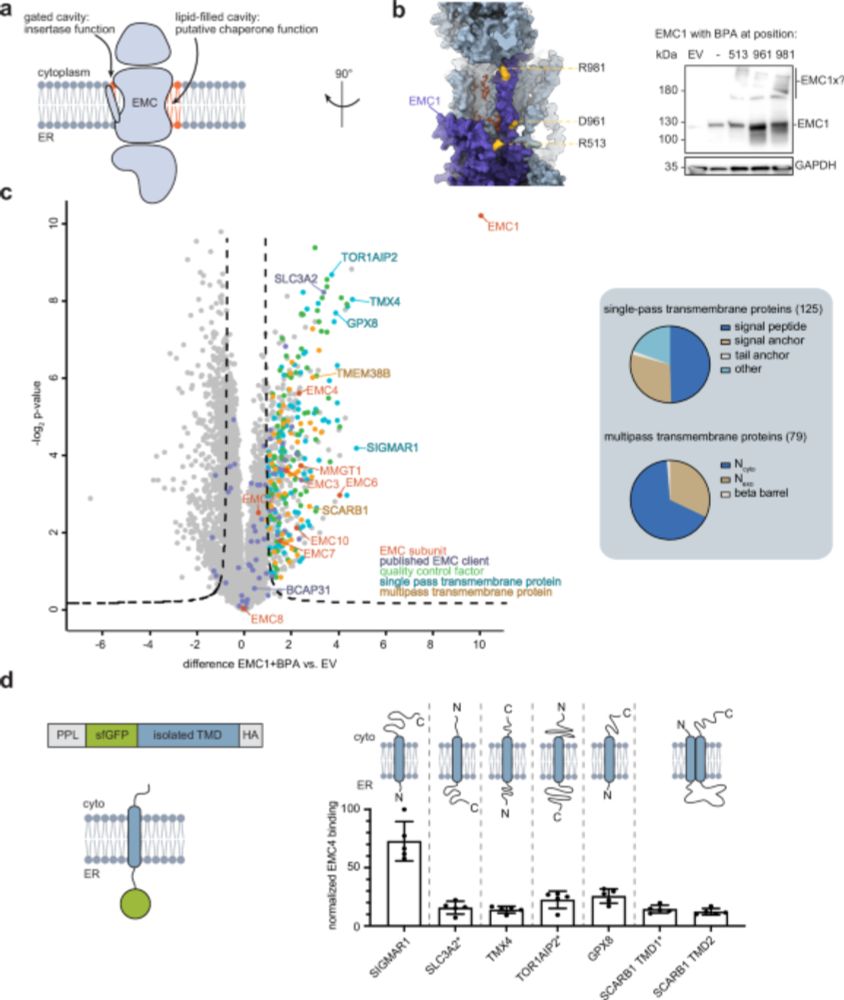
Formation & function of #MembranelessOrganelles! #CryoET structures of #proteasome storage granules inside cells!
Read our paper @cp-cell.bsky.social!
❕Publication: doi.org/10.1016/j.ce...
❕Press Release: www.biochem.mpg.de/en/pressroom
@uoftmedicine.bsky.social
@erc.europa.eu #UPSmeetMet
28.01.2026 16:39 —
👍 68
🔁 26
💬 0
📌 2
We’re excited to share our latest preprint on the mechanism of excised linear intron stabilization in yeast! This work was led by PhD student @glennli.bsky.social and was a wonderful collaboration with @maxewilkinson.bsky.social. Link: www.biorxiv.org/content/10.6... (1/4)
23.01.2026 16:14 —
👍 58
🔁 25
💬 1
📌 2

Pour a glass of champagne AND red Bordeaux—Our newest work with GSK @scripps.edu is out in @nature.com! Here we describe SB-405483, the first allosteric CRBN ligand which potentiates neosubstrate degradation. Congrats Vanessa and all authors! 🍷💫💐 www.nature.com/articles/s41...
21.01.2026 16:29 —
👍 50
🔁 11
💬 1
📌 4
This is also a good occasion to highlight that we are looking for post-doctoral researchers that are interested in understanding the fundamental mechanisms of transcription, DNA replication, and chromatin.
13.01.2026 20:37 —
👍 9
🔁 3
💬 0
📌 0
Check out the newest preprint from the Schulman lab 🎊🎉
Super cool mechanism of how a metabolite regulates the stability of its own metabolizing enzyme!
Alina did it all for this project CRISPR screen ✂️, biochemistry 🧪, and cryo-EM ❄️🔬.
Congrats!
13.01.2026 14:56 —
👍 13
🔁 5
💬 0
📌 0
What happens when E3 Ubiquitin Ligase and RNA Enthusiasts team up!
Massive congrats to this transatlantic collaboration between @jakobfarnung.bsky.social from Schulman Lab and @elenaslo.bsky.social from @bartellab.bsky.social
Have a read!
06.01.2026 15:50 —
👍 9
🔁 2
💬 0
📌 0
The E3 ubiquitin ligase mechanism specifying target-directed microRNA degradation www.biorxiv.org/content/10.64898/2026.01.05.697729v1 #cryoEM
06.01.2026 12:00 —
👍 6
🔁 3
💬 0
📌 0
Look at this super exciting E3, recognizing the specific RNA-bound state of it’s target protein alone!
Massive Congrats @jakobfarnung.bsky.social and collaborators from @bartellab.bsky.social
06.01.2026 09:02 —
👍 6
🔁 1
💬 0
📌 0
This work has been an amazing✈️ transatlantic 🌎 collaboration. A special shout-out goes to @elenaslo.bsky.social , establishing new assays, exchanging hundreds of emails and just being and amazing amazing collaborator while the Schulman lab dipped its toes into the waters of RNA biology. (5/5)
06.01.2026 08:04 —
👍 7
🔁 0
💬 0
📌 0
For all the E3 ligase-afficionados, we could show how ZSWIM8 recruits CUL3 despite also interacting with ELOB/C. By positively selecting for CUL3 and negatively selecting against other cullins, ZSWIM8 establishes a distinct family of E3 ligases. (4/5)
06.01.2026 08:04 —
👍 0
🔁 0
💬 1
📌 0

Why does ZSWIM8 use such an intricate recognition mode? Trigger-bound AGO complexes are quite sparse in the cell compared to the thousands of other AGO complexes that shouldn’t be targeted by ZSWIM8. By recognizing RNA and protein conformation ZSWIM8 achieves exquisite selectivity. (3/5)
06.01.2026 08:04 —
👍 1
🔁 0
💬 1
📌 1

Using biochemistry 🧪, cryo-EM 🔬 and cell biology 🧫, we show how pairing of trigger RNAs to microRNAs induces a unique AGO-microRNA-trigger RNA conformation. Dimeric ZSWIM8 engulfs this complex recognizing both RNA- and protein conformation that specify AGO for ubiquitin-mediated degradation (2/5).
06.01.2026 08:04 —
👍 5
🔁 0
💬 1
📌 0
When RNA Degradation 🤝 meets 🤝 Protein Degradation! tinyurl.com/E3TDMD In a collaboration of @bartellab.bsky.social and Schulman lab, we show that, in target-directed microRNA degradation (TDMD), 2-RNA-factors recruit an E3 ligase and induce the degradation of not only a protein but also RNA (1/5).
06.01.2026 08:04 —
👍 117
🔁 50
💬 1
📌 4

New research group leader for functional genomics
Molecular biologist Matthias Muhar becomes part of the MPI-CBG faculty
New research group leader @matthiasmuhar.bsky.social joins @mpi-cbg.de! 🥳 With his group "Functional genomics of proteome remodeling,” Matthias wants to pursue high-throughput genetic studies to understand how protein turnover is regulated. Welcome, Matthias! www.mpi-cbg.de/news-outreac...
11.12.2025 14:15 —
👍 43
🔁 15
💬 0
📌 1
Cysteine availability tunes ubiquitin signaling via inverse stability of LRRC58 E3 ligase and its substrate CDO1 https://www.biorxiv.org/content/10.1101/2025.11.14.688510v1
15.11.2025 02:45 —
👍 8
🔁 6
💬 0
📌 0
Have a look at this Tools of the Trade Article I wrote for @natrevmcb.nature.com on decoding #ubiquitin signals inside cells using UbiREAD!
Many thanks to @lisaheinke.bsky.social for the opportunity to write this TotT!
04.11.2025 10:04 —
👍 24
🔁 12
💬 0
📌 1
🧬 Transcription elongation by RNA polymerase II relies on a web of elongation factors. Our new work shows how IWS1 acts as a modular scaffold to stabilize & stimulate elongation. Fantastic work by Della Syau! www.biorxiv.org/content/10.1...
29.08.2025 10:14 —
👍 57
🔁 23
💬 1
📌 1

Excited about #ubiquitin and #TPD ?
Then join us tomorrow for the latest instalment of the @danafarber.bsky.social Targeted Protein Degradation Webinar where Kimberly Stegmaier and I will present our latest work.
dfci.zoom.us/webinar/regi...
25.06.2025 21:08 —
👍 13
🔁 5
💬 0
📌 0

The 2025 Freeman Hrabowski Scholars | HHMI
Freeman Hrabowski Scholars are outstanding early career faculty who have the potential to become leaders in their research fields.
Congratulations to our own Lucas Farnung @lucas.farnunglab.com who has been named as one of 30 Freeman Hrabowski Scholars for 2025 by the Howard Hughes Medical Institute (HHMI). So well deserved deserved !! www.hhmi.org/programs/fre...
18.06.2025 15:25 —
👍 28
🔁 6
💬 0
📌 0

Schulman lab is ready for the GRK2243 Symposium: Understanding ubiquitination: from molecular mechanisms to disease in würzburg
#wUeBI2025 @grk2243.bsky.social
@jakobfarnung.bsky.social @samuelmaiwald.bsky.social @hannahbkmpr.bsky.social
02.06.2025 10:15 —
👍 27
🔁 6
💬 2
📌 0
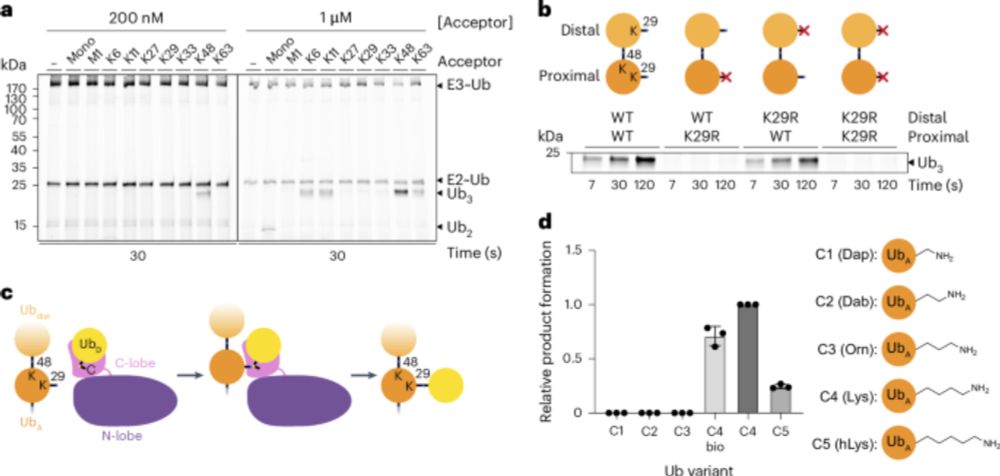
Super cool story on branched UB chain formation. Congrats Sam!
26.05.2025 09:48 —
👍 10
🔁 3
💬 1
📌 0

🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
06.05.2025 15:08 —
👍 103
🔁 24
💬 4
📌 3