Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇
24.09.2025 21:45 — 👍 102 🔁 50 💬 7 📌 5
4/
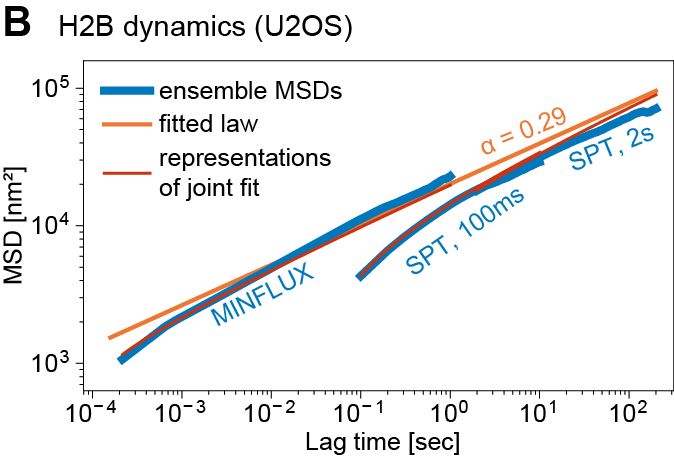
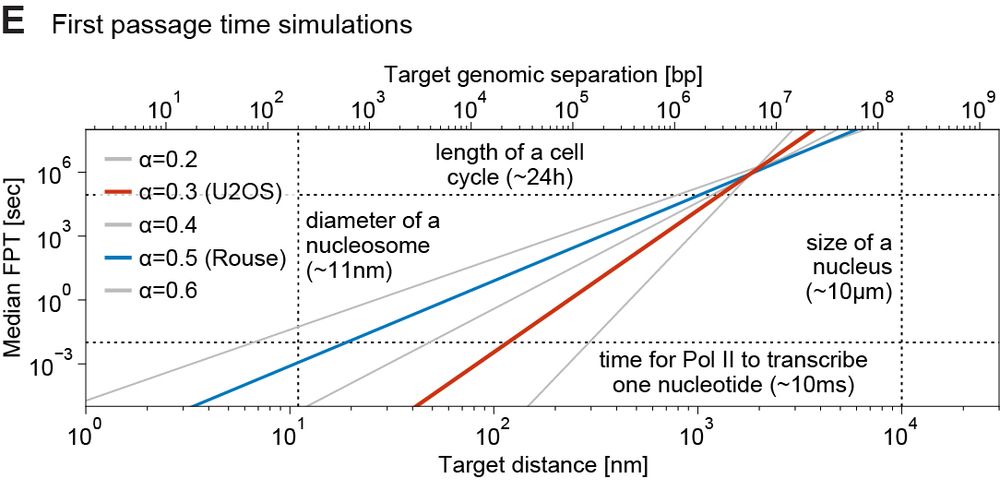
However, as distance increases search times become progressively more challenging. At ~1µm (1-3 Mb), EP can take ~5-24 h to randomly find each other – potentially too long for a cell cycle → distant EP need help (e.g. loop extrusion)
14.05.2025 17:58 — 👍 4 🔁 0 💬 1 📌 0
3/
We ran simulations to estimate EP search times based on 1) measured subdiffusion, 2) physical/genomic distance. Within 300nm (~<200 kb), a gene can find 1000s of enhancers in minutes. Such encounters can’t be selective → EP selectivity needs other mechanisms, e.g. biochemical
14.05.2025 17:58 — 👍 3 🔁 1 💬 1 📌 0
2/
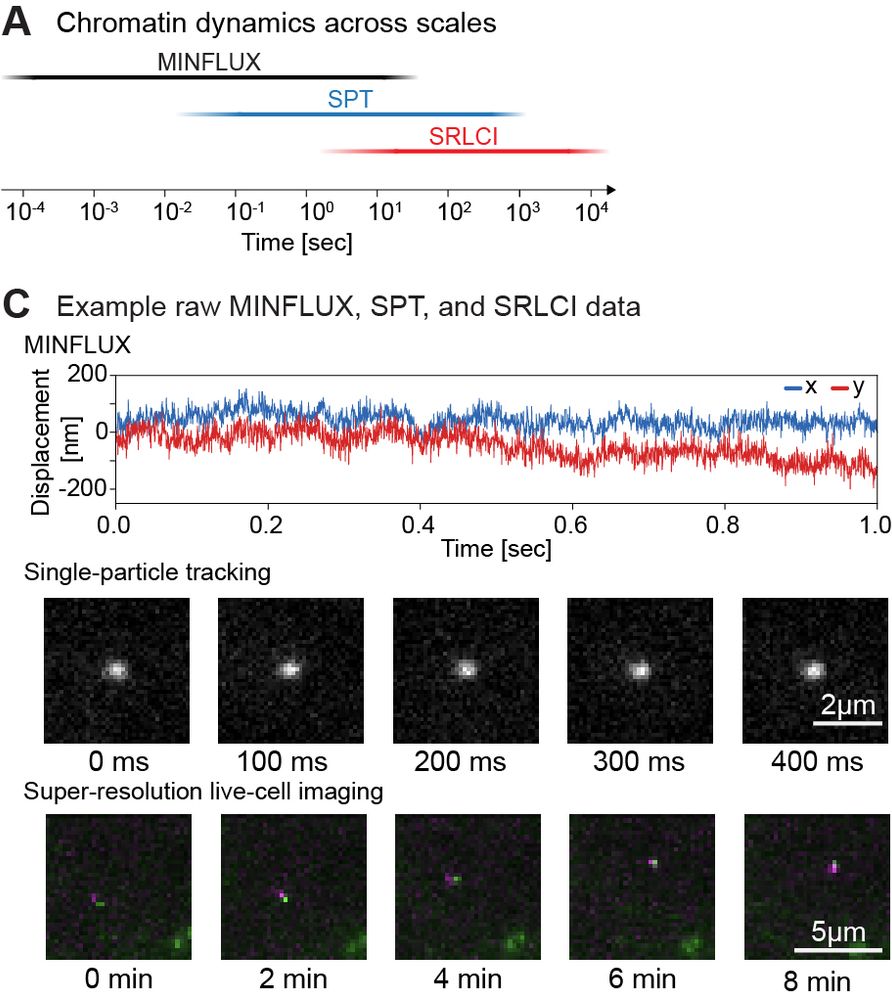
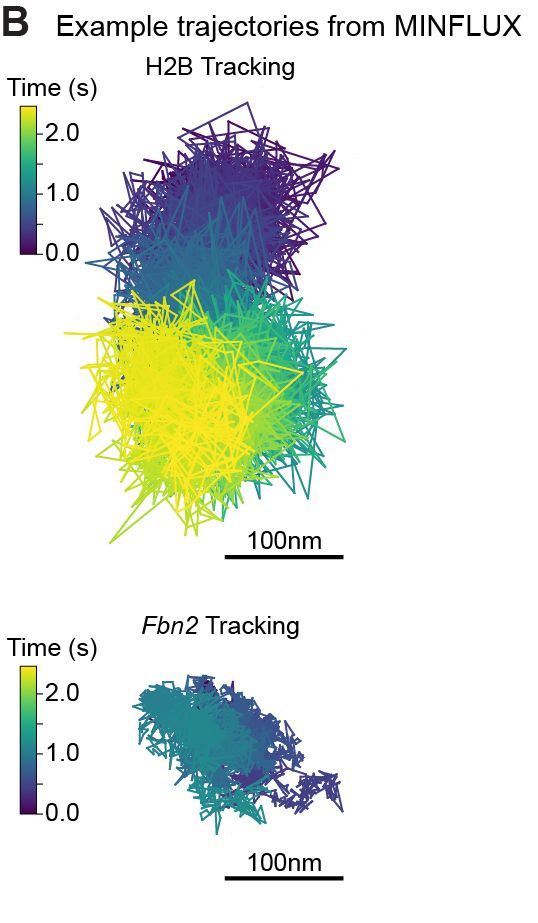
Such fast and long chromatin tracking is key to understanding how distal genomic elements (e.g. enhancers and promoters – EP) find each other. We found chromatin is highly subdiffusive (α ~ 0.3) -> loci are great at exploring the neighborhoods but rarely reach distant regions
14.05.2025 17:58 — 👍 4 🔁 0 💬 1 📌 0
1/
Super excited to share my first preprint from @andersshansen.bsky.social Lab! We used MINFLUX to track chromatin (H2B-Halo and Fbn2 locus) at an unprecedented 200 μs, then combined it with SPT to span μs-minutes (H2B) or SPT & Super-Res Live-Cell Imaging (SRLCI) to span μs-hours (Fbn2)
14.05.2025 17:58 — 👍 28 🔁 11 💬 1 📌 4
Mentor, scientist & engineer. Having fun in @slavovlab.bsky.social and Parallel Squared Technology Institute @parallelsq.bsky.social with biology & single-cell proteomics.
https://nikolai.slavovlab.net
PhD Student in Luca Giorgetti's Lab (@lucagiorgetti.bsky.social) - Genome architecture - Microscopy - Image analysis - Biophysics
Biophysicist & computational biologist | Genome organization, chromatin modeling
Postdoc at Institut Curie, Paris (Leonid Mirny group) | Formerly at IMBA, Vienna (Anton Goloborodko group)
Postdoc at FMI - studying chromosome folding and dynamics in relation to transcriptional regulation, using live-cell imaging and polymer modeling.
Director, Dept. of Mechanistic Cell Biology, Max Planck Institute of Molecular Physiology, Dortmund (DE). Opinions my own
PhD Student with @marcbuhler.bsky.social at @fmiscience.bsky.social
Master's in Heidelberg, thesis with @karsten-rippe.bsky.social
Editorial assistant at the @nightsciencepod.bsky.social
gene regulation, chromatin, nuclear organisation, synthetic biology
Physics at Ulm University | live-cell/organism single-molecule microscopy | super-resolution microscopy | gene regulation | chromatin architecture | embryo development
Postdoctoral researcher in the Sebe-Pedros and Marti-Renom Labs at CRG. Transposable elements enthusiast, passionate about piRNAs, 3D genomes, and Star Trek 🖖
Our laboratory seeks to understand how chromosome structure relates to genome functions
Associate Professor & Docent @umeauniversitet.bsky.social
Wallenberg Fellow in Molecular Medicine at WCMM Umeå
Postdoc @embl.org
PhD @cniostopcancer.bsky.social
3D genomics , epigenomics, gene regulation, enhancer, cancer, glioblastoma, cancer neuroscience
Ph. D. Student in Maeshima lab in NIG @kazu-maeshima.bsky.social
Biophysics, Chromatin / Kyoto Uni. → NIG
We study transcriptional regulation and chromosome folding using an interdisciplinary approach combining wet- and dry-lab methods.
https://giorgettilab.org
@fmiscience.bsky.social
The mission of the Center for Physical Genomics and Engineering at Northwestern University’s McCormick School of Engineering is to create new strategies for the treatment of disease and the reversible manipulation of living systems
Group leader @mpi-mg, Berlin
Chromatin tracing, epigenetics, development. Equity and advocacy in academia.
PhD student at MIT Physics. Interested in statistical physics, gene regulation, and fundamental questions in biology.
Postdoctoral researcher at TU Delft.
Trying to sequence proteins with #nanopore
News from the Sternberg lab at Columbia University, Howard Hughes Medical Institute.
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
Single Molecule Tracking. Postdoc at Tjian-Darzacq lab, UC Berkeley. Biophysics PhD with Biteen lab UMICH.





