
Cilia alert! Stoked to have the new paper from Juyeon Hong about a new domain at the extreme distal tip of motile cilia! (The EDT, y'all!) It's out now @natcomms.nature.com.
Check it out!
#cilia
www.nature.com/articles/s41...
@builab.bsky.social

Cilia alert! Stoked to have the new paper from Juyeon Hong about a new domain at the extreme distal tip of motile cilia! (The EDT, y'all!) It's out now @natcomms.nature.com.
Check it out!
#cilia
www.nature.com/articles/s41...

The entire video list with Installation, Build Cilia, Centriole & 2D template
www.youtube.com/playlist?lis...

CiliaBuilder Video Tutorial
www.youtube.com/watch?v=SJHo...
If your installation didn't work a few days ago, try again. I've fixed a few things, and let me know if it still doesn't work for you.
I just noticed that I did not put the proper dependency. Now, if you download version 1.0.6, it will install the dependency properly to run.
24.12.2025 15:54 — 👍 0 🔁 0 💬 0 📌 0I am planning to do the next release with ArtiaX style cilia in the near future :) You probably will like that a lot more.
23.12.2025 20:13 — 👍 2 🔁 0 💬 0 📌 0This was built in ~10 days of vibe-coding with Claude & Gemini. Now officially on holiday 😌
Video tutorial coming after the holidays



Tired of drawing cilia or centrioles for slides and papers? I feel you.
Check out CiliaBuilder for ChimeraX → cxtoolshed.rbvi.ucsf.edu/apps/chimera...
Build custom 3D cilia & centriole models (tips + primary cilia included).
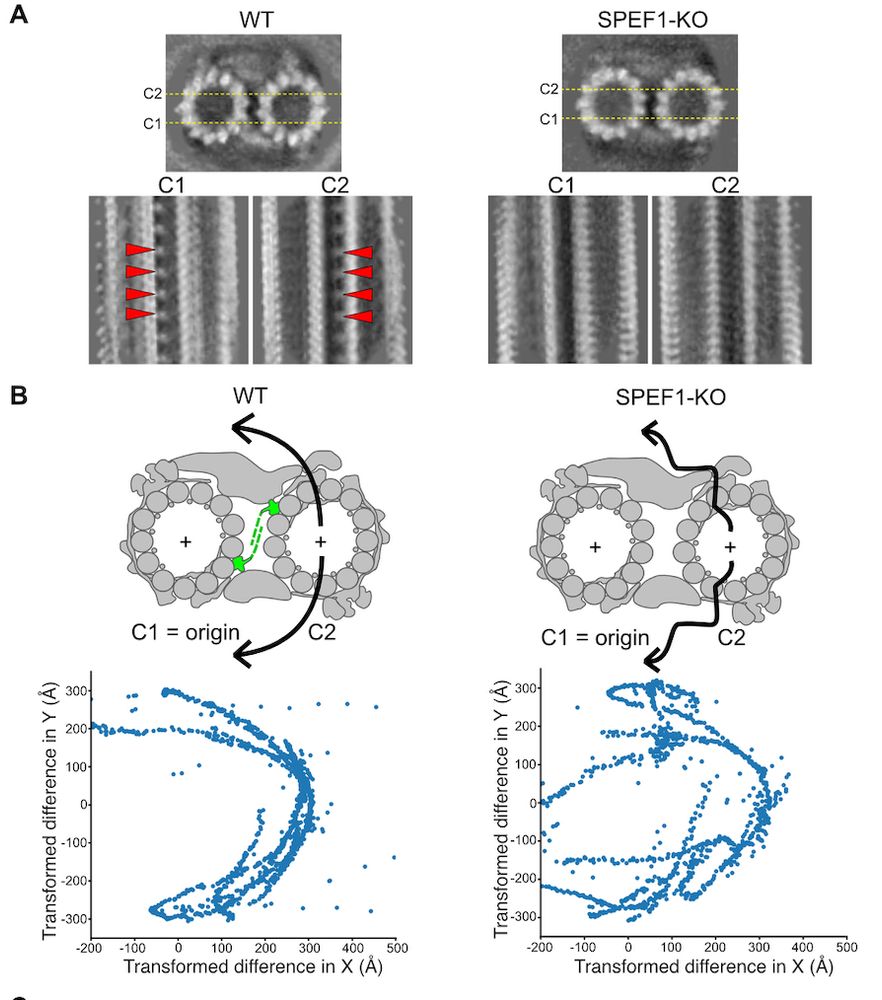
We also analysed SPEF1-KO cilia by cryo-ET to show that SPEF1 restricts C1 and C2 microtubule movement.
11.07.2025 19:41 — 👍 0 🔁 0 💬 0 📌 0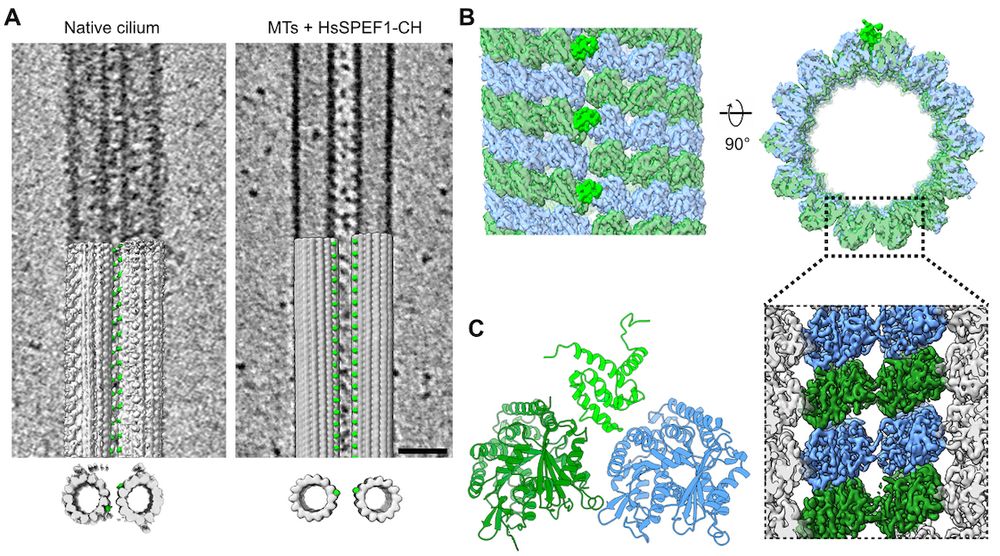
In the final version, we included a lot more data and analysis compared to the previous biorxiv version. We achieved a 4.6 Angstrom resolution of the microtubule by subtomogram averaging of MT-SPEF1 complexes.
11.07.2025 19:41 — 👍 2 🔁 0 💬 1 📌 0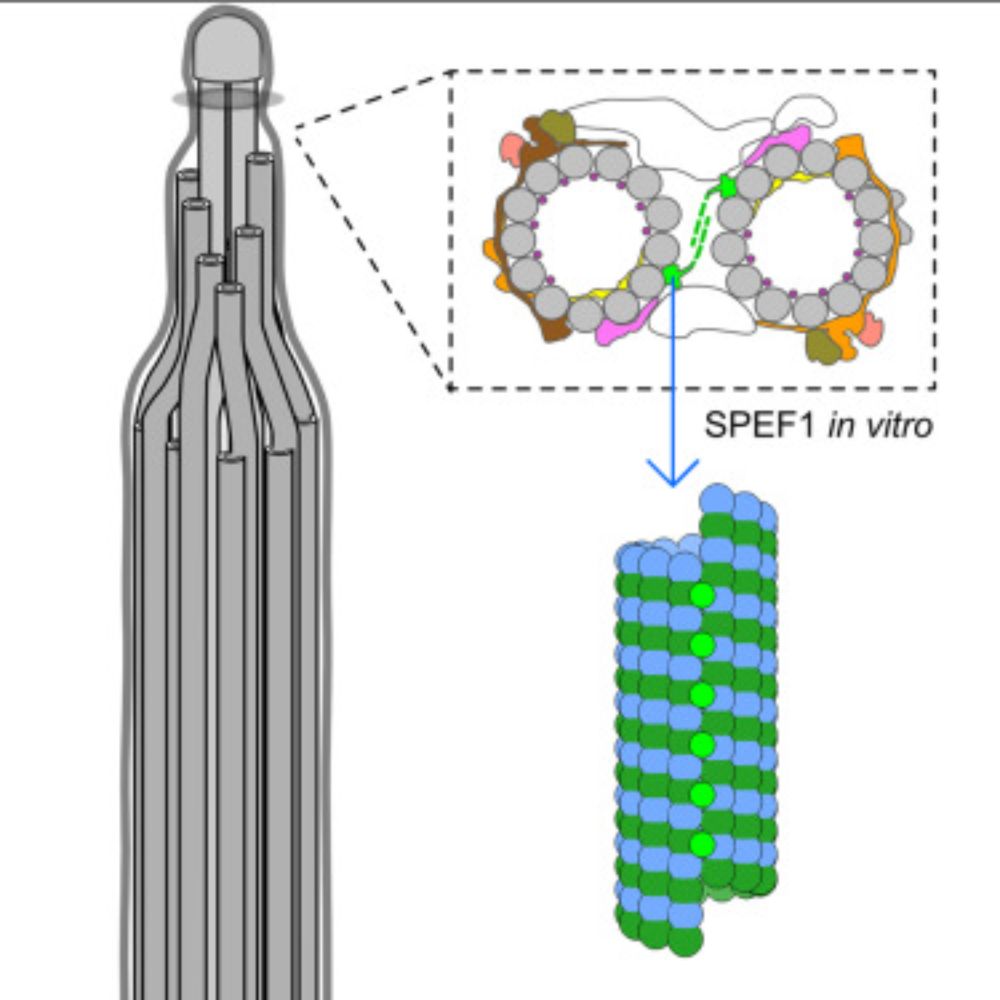
Our newest paper is now online in Current Biology. In short, we identified 7 proteins in the tip of the cilia and found SPEF1 is a seam-binding protein and important for cilia stability. Great work from my postdoc Thibault Legal and everyone involved!
www.sciencedirect.com/science/arti...
This seems great. I want to working on our picking using AI in the summer. Perhaps, I can ask you for an installation and usage guide if I have problems.
08.03.2025 23:28 — 👍 0 🔁 0 💬 1 📌 0I hated Amira with the $$. Need to try DragonFly but most of our processing is done on the cluster so anything using command line is a big advantage. Perhaps, my summer project :)
06.03.2025 05:21 — 👍 1 🔁 0 💬 1 📌 0interesting I want to use dragonfly for segmentation but can we run dragonfly in command line for batch mode? Do you have a script to cluster and turn dragon fly into filament light?
05.03.2025 04:32 — 👍 1 🔁 0 💬 1 📌 0Very nice preprint from the Wallingford lab. Seems like there is a common theme of tip proteins as these proteins seem to be in the tip of the ciliate cilia too.
05.03.2025 04:27 — 👍 4 🔁 1 💬 1 📌 0Thanks. I found template matching is too slow for my need as I can manual pick microtubules much faster and more accurate. These tryout takes time so I 'd love to see people's experiences.
05.03.2025 02:34 — 👍 1 🔁 0 💬 1 📌 0#TeamTomo What is the open-source and good cryoET deep-picking program?
26.02.2025 15:41 — 👍 2 🔁 0 💬 1 📌 0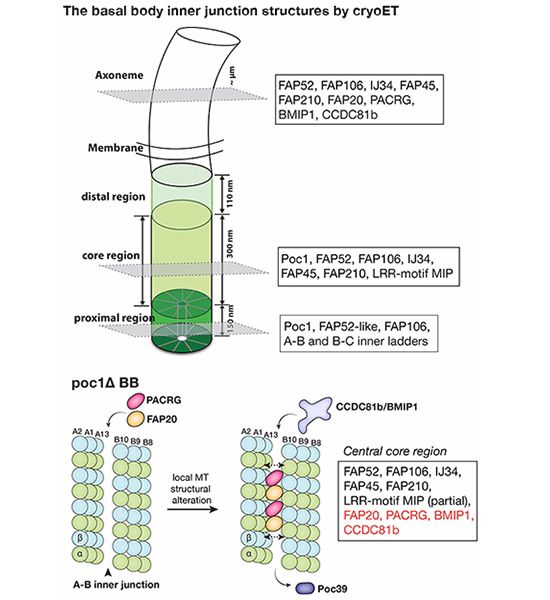
Cryo-electron tomography of Tetrahymena #cilia by Sam Li, David Agard, Mark Winey provides insights into the intricate spatial organization of basal body inner junctions, compromised by loss of Poc1
www.embopress.org/doi/full/10....

We have posted our new #preprint regarding the track recognition mechanism of dynein-2 that works in #cilia on bioRxiv @biorxivpreprint.bsky.social!!
www.biorxiv.org/content/10.1...
1/8 🧵

Excited to share our work on bioRxiv before my thesis defense! Together with my undergraduate mentee Diego, postdocs Wan and Jun, and my advisor Kai, we identified a novel dynein heavy chain subfamily, DNAHX, from sea urchin sperm axoneme.
Check out the details here: www.biorxiv.org/content/10.1...
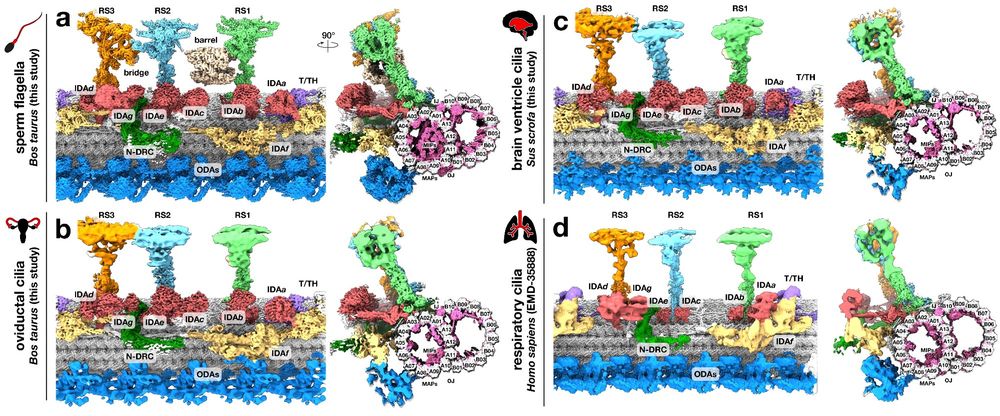
Best way to start 2025! Thrilled to share our Nature paper about the structures of axonemal components from sperm flagella and from epithelial cilia of the oviduct and brain ventricles. Great collaboration with Tzviya Zeev-Ben-Mordehai and @alanbrownhms.bsky.social
www.nature.com/articles/s41...
Amazing work. It is so useful for us working with Tetrahymena and for cilia in general.
17.12.2024 23:13 — 👍 1 🔁 0 💬 0 📌 0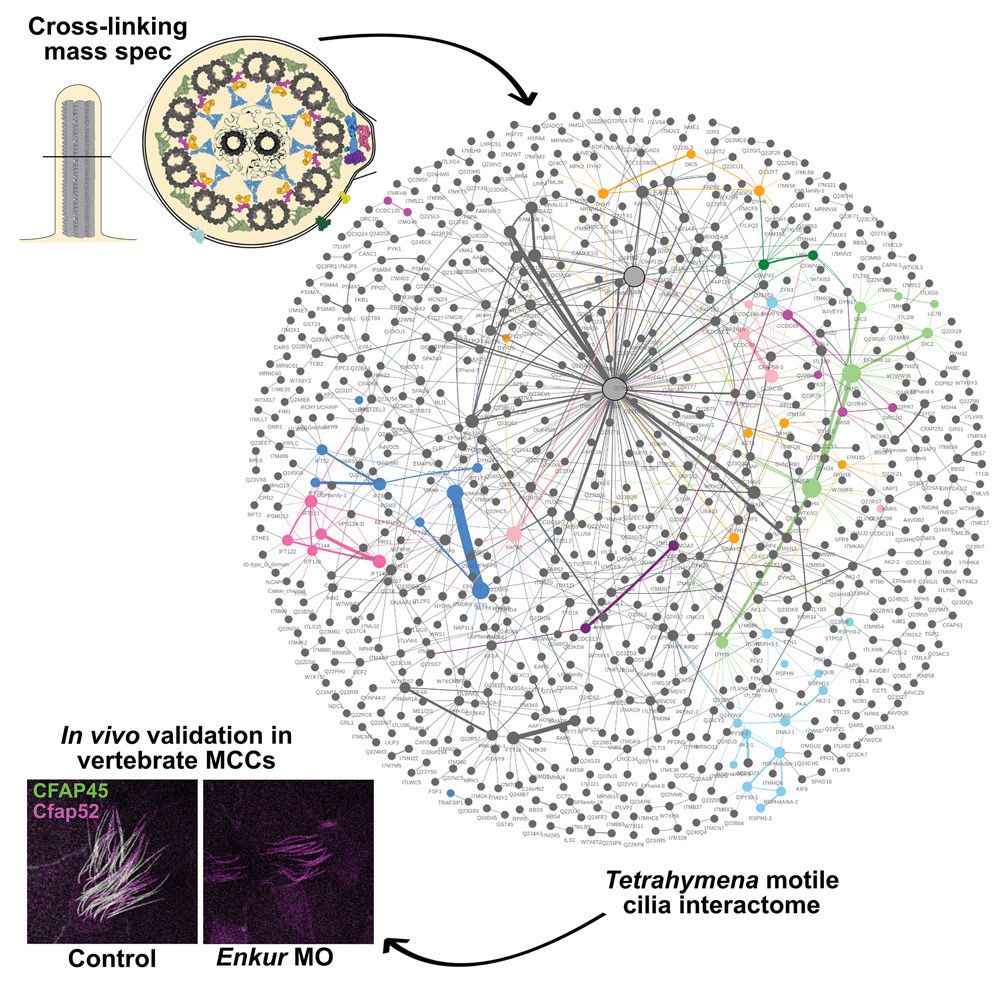
I am excited to share some of my PhD work on the motile cilia interactome revealed by XL/MS with @jbwallingford.bsky.social @edwardmarcotte.bsky.social Ophelia Papoulas, Chanjae Lee, David Taylor, and @builab.bsky.social www.cell.com/developmenta...
16.12.2024 14:44 — 👍 67 🔁 18 💬 4 📌 5ChimeraX 1.9 has been released and is available now on our website
13.12.2024 17:49 — 👍 27 🔁 8 💬 0 📌 0Thanks. Warp on Linux is awesome. This is my Twitter import so it is very old tweet
05.12.2024 14:10 — 👍 0 🔁 0 💬 2 📌 0My last post on Twitter before.
04.12.2024 17:06 — 👍 1 🔁 0 💬 0 📌 0Proudly present our new preprint on my first post here. We performed SPA on the ciliary tip and identify 7 proteins. Among them, we found the conserved protein SPEF1 at the microtubule seam. In vitro assays show that SPEF1 binds to the seam and stablizes microtubules.
www.biorxiv.org/content/10.1...

The Bui lab Twitter will stop and we are migrating to Mastodon. Anyway, the last post is to thank the @Beck_Laboratory and @jankosinski @aobarska for the new wall art decoration. NPC rules :
19.12.2022 02:33 — 👍 2 🔁 0 💬 1 📌 0Our paper on the T- and N-pilus structure with the @ChristianBaron and @NatalieZeytuni labs is finally online. Great work from Amro Jaafar and @TheRealCSBlack and others.
authors.elsevier.com/a/1gElW3SNvc2%…
Taking forever but finally here. We identified close to 40 MIPs in Tetrahymena doublet. Specifically, the MIPs at the outer junction (CFAP77, OJ2 & OJ3). @dynein_awk @TheRealCSBlack @ahmad__khalifa
03.10.2022 13:15 — 👍 2 🔁 0 💬 0 📌 0Just update the subtomo2Chimera withto visualize subtomo from Dynamo. Also, small fixes for relionsubtomo2ChimeraX. now you can also using stl file for visualization for more efficient memory #TeamTomo
github.com/builab/subtomo… dynamotable2ChimeraX.py