Shout out to wonderful collaborator @briankalishMD and funding from @hhmi_science @CSHL @NIMHgov @McKnightFdn Rita Allen Foundation
04.02.2026 16:33 — 👍 0 🔁 0 💬 0 📌 0
This study underscores the importance of studying autism not just from a neurological perspective but in the context of immunology as well.
04.02.2026 16:32 — 👍 0 🔁 0 💬 1 📌 0
Follow up experiments directly testing this hypothesis and extending our findings to human biology are underway.
04.02.2026 16:32 — 👍 0 🔁 0 💬 1 📌 0
We hypothesize that male embryos express unique factors, i.e. antigens that are non-self to the female dam, that, when combined with a loss of placental immunosuppression due to maternal infection, derails fetal development in males.
04.02.2026 16:32 — 👍 0 🔁 0 💬 1 📌 0
We hypothesize that male embryos express unique factors, i.e. antigens that are non-self to the female dam, that, when combined with a loss of placental immunosuppression due to maternal infection, derails fetal development in males.
04.02.2026 16:32 — 👍 0 🔁 0 💬 0 📌 0
What we think is most exciting: out of hundreds of embryos analyzed, only males exhibited these phenotypes. Female embryos and placentas were protected in every single case.
04.02.2026 16:31 — 👍 0 🔁 0 💬 2 📌 0
Innate immune cells and cytokines accumulate within the amniotic fluid, with one of these cytokines, IL6, being necessary for the deficits to emerge.
04.02.2026 16:31 — 👍 0 🔁 0 💬 1 📌 0
The placentas of affected male embryos exhibit structural deterioration near the region where maternal and fetal compartments meet.
04.02.2026 16:31 — 👍 0 🔁 0 💬 1 📌 0
Concurrently, placental cells called spongiotrophoblasts decrease ECM and hormone expression following MIA, leading to a loss of immunosuppression at the maternal-fetal interface.
04.02.2026 16:30 — 👍 0 🔁 0 💬 1 📌 0
These abnormalities coincide with the induction of a pro-inflammatory placental microenvironment, with both maternally and fatally derived immune cells mounting inflammatory responses.
04.02.2026 16:30 — 👍 0 🔁 0 💬 1 📌 0
Using a mouse model, we find that only 24 hours of maternal immune activation (MIA) during pregnancy leads to robust developmental deficits in 30% of embryos.
04.02.2026 16:29 — 👍 0 🔁 0 💬 1 📌 0
Two major gaps in knowledge: 1. What inflammatory triggers derail fetal brain development? 2. Why are some individuals susceptible to these triggers while others are not?
04.02.2026 16:29 — 👍 0 🔁 0 💬 1 📌 0
While genetic factors contribute to autism, environmental factors such as prenatal inflammation can also play a key role. For example, maternal infections during pregnancy increase autism risk in the offspring.
04.02.2026 16:29 — 👍 0 🔁 0 💬 1 📌 0
Excited to share new work from the Cheadle Lab investigating the inflammatory basis of autism spectrum disorder (tweetorial below)
biorxiv.org/content/10.648…
04.02.2026 16:28 — 👍 0 🔁 0 💬 1 📌 0
So glad we had the chance to contribute to this beautiful study on neuronal function in breast cancer. Fantastic collaboration w @jborniger.bsky.social and @corina-amor.bsky.social
15.12.2025 16:51 — 👍 2 🔁 0 💬 0 📌 0
Thank you!
04.11.2025 00:02 — 👍 0 🔁 0 💬 0 📌 0
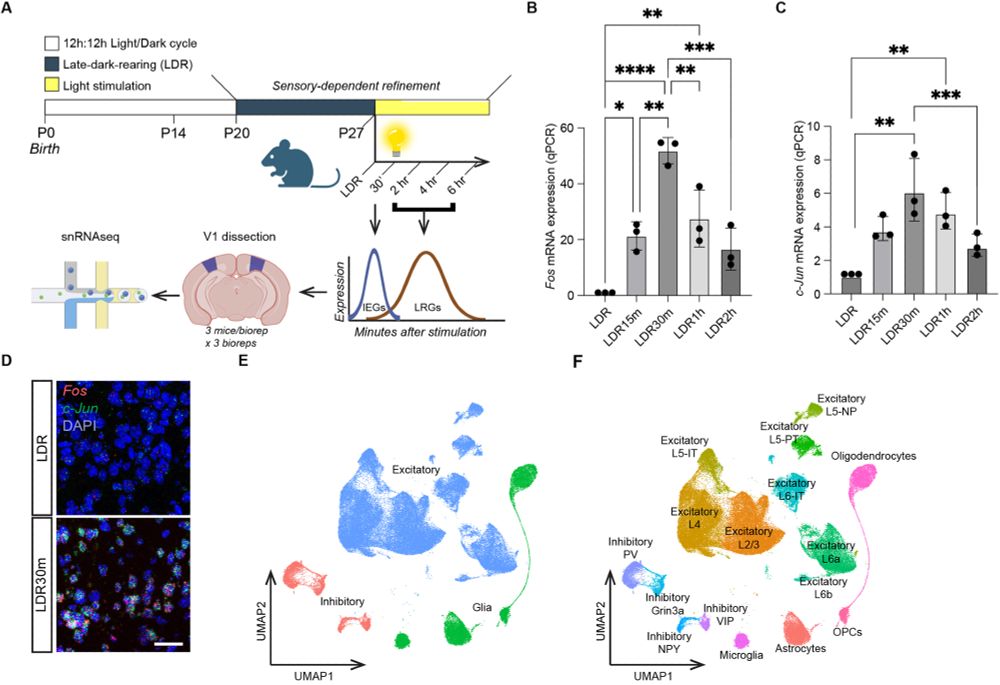
Figure 1 - Experimental design and introduction to the single-nucleus RNA sequencing dataset. (A) Schematic illustrating the late dark-rearing (LDR) paradigm and the workflow of the single-nucleus RNA sequencing (snRNAseq) experiments. (B) Quantification of Fos mRNA expression in sensory deprived (LDR) mice and in mice acutely exposed to light for between 15 min and 2 h, with stimulation timepoints labeled as follows: LDR15m (15 min of light), LDR30m (30 min), LDR1h (1 h) and LDR2h (2 h). Fos expression assessed by qPCR and normalized to Gapdh. Values plotted are additionally normalized to the LDR condition. (C) qPCR quantification of Jun mRNA expression (normalized to Gapdh) in V1 across all timepoints. Data obtained by qPCR and values plotted are normalized to LDR. In B and C, data are mean±s.e.m. n=3 mice per condition. One-way ANOVA followed by Tukey's post hoc test: *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. (D) Example confocal images of V1 in sections from a sensory deprived mouse (LDR) and a mouse re-exposed to light for 30 min (LDR30m). Fos mRNA (red), Jun mRNA (green) and DAPI (blue). Scale bar: 44 µm. (E) UMAP plot illustrating the 118,529 nuclei in the dataset categorized by general cell class: excitatory neurons (blue), inhibitory neurons (pink) and glia (green). (F) UMAP plot with all 16 clusters colored and labeled by cell type.
A single-cell transcriptomic atlas of sensory-dependent gene expression in developing mouse visual cortex
Read this this #LifelongDevSI #OA Techniques and Resources Article by Andre M. Xavier, Qianyu Lin, Chris J. Kang, Lucas Cheadle (@cheadlelab.bsky.social)
doi.org/10.1242/dev....
03.11.2025 15:59 — 👍 4 🔁 1 💬 0 📌 1
Thrilled to welcome the new FHS cohort… including these newly selected McKnight Scholars!
20.06.2025 18:39 — 👍 5 🔁 1 💬 0 📌 0
That’s my plan!
31.05.2025 16:47 — 👍 1 🔁 0 💬 0 📌 0
Wish I had known you were around!
31.05.2025 16:37 — 👍 0 🔁 0 💬 1 📌 0
Love this
10.05.2025 17:32 — 👍 1 🔁 0 💬 0 📌 0
Hosting Dr. Kaf Dzirasa @hhmi.org for a seminar at CSHL and getting to spend time w/fellow FHS @ishmailsaboor.bsky.social in the process has made this week a highlight of my year. Feeling seriously inspired today.
24.04.2025 18:23 — 👍 4 🔁 0 💬 0 📌 0
Congrats!
20.03.2025 15:35 — 👍 1 🔁 0 💬 0 📌 0
Awesome to see this!
11.03.2025 20:18 — 👍 1 🔁 0 💬 1 📌 0
I’m curious whether the men were mentored to say “no” while the women were mentored to say “yes”. My mentors have always taught me to embrace saying “no” but maybe this advice differs by gender?
08.03.2025 22:15 — 👍 1 🔁 0 💬 0 📌 0
Delighted to share the published version of this work from brilliant #CancerNeuroscience post-doc Tara Barron detailing GABAergic neuron-to-glioma synapses in #DIPG/#DMG. 🧵
Open access link:
www.nature.com/articles/s41...
19.02.2025 21:41 — 👍 74 🔁 23 💬 2 📌 0
Dear friends and colleagues: nice to see you here where the skies are bluer. Follow me for updates on our research into neuro-immune interactions in brain development, function, and disease. @cshlnews.bsky.social @hhmi.bsky.social
27.01.2025 16:47 — 👍 1 🔁 0 💬 0 📌 0
Professor of Neuroscience. Studying neural development, regeneration, and control of innate behaviors at Johns Hopkins.
Inupiaq, scientist of host-microbe interactions, mom, runner, news junkie, not necessarily in that order. I speak for myself. She/her. Only reskeets posts with alt text. Unapologetically typo prone, doesn't use AI.
Indigenous (Diné/Navajo) geneticist and bioethicist | Indigenous-led research and data innovation
Non-profit research and educational institution at the forefront of molecular biology, cancer, neuroscience, plant biology, and genetics.
https://www.cshl.edu/
Entrepreneur
Costplusdrugs.com
Focused on innate immune mechanisms governing pathogenesis of disease and on NK-cell based therapies for disease. Opinions expressed are Dr. Waggoner's alone and not on behalf of lab staff or employer.
Advanced Light Microscopy facility based at Kings College London, Denmark Hill Campus
Proudly a Nikon Centre of Excellence.
Neuropharmacology Lab at a major R1 in the south. We study mechanisms of and drug discovery for alcohol-induced neurodegeneration.
neural stem cells | adult neurogenesis | microglia | adolescent development | aging
Posts are our opinion.
Official Journal of Neuroimmunology News Twitter. Latest articles on Immune & Brain interface research. Fellow @JNI_Journal for latest research news
Senior Scientist, Associate Professor & Program Director @ OHRI & uOttawa 🇨🇦. Fascinated by vascular & metabolic causes of neurodevelopmental disorders, SciArt and the night sky 🔬 🎨 🔭 Follow for 🧪 news and more! Look for #cerebrovascular
Brain development enthusiast 🧠 @ University of Manchester
fongkuanwonglab.com
Waitress turned Congresswoman for the Bronx and Queens. Grassroots elected, small-dollar supported. A better world is possible.
ocasiocortez.com
Neuroscientist. Neuro-Oncologist. Mom.
Associate Prof in cancer neuroscience at CSHL; amateur astrophotographer; views are mine alone
https://www.cshl.edu/research/faculty-staff/jeremy-c-borniger/ 🇵🇸
Neuroscientist and mom. Interested in brain development, early life stress, epigenetics, cats, mountains, donuts, and lots of coffee.
www.PenaLab.org
Asst Prof | Harvard
Freeman Hrabowski Scholar | HHMI
Founder | Leading Edge @leadingedgeprogram.bsky.social
My lab studies menstruation and menstrual disorders.
www.mckinleylab.org
Opinions are my own and do not reflect those of my employer.
Wellcome Career Development fellow investigating how the immune system modulates behaviour during chronic challenges - https://www.quintanalab.com
Translational MD-scientist, mom, 🎶 | Bklyn born + bred
Pathogen/microbe-host interactions + 🧠outcomes across the life course | gut-immune-🧠 axis
#ASD #ADHD #PANS/#PANDAS #OCD affective disorders/#TRD Alzheimer’s #MECFS #LongCOVID
All posts = my views
Fan of microglia, macrophages, glial-neuronal and brain-immune interactions (both innate & adaptive); flirted with oligodendrocytes, astrocytes and occasionally even neurons.
EIC Journal of Neuroinflammation
Sci-Fi and SDCC (Comicon)
opinions are my own


