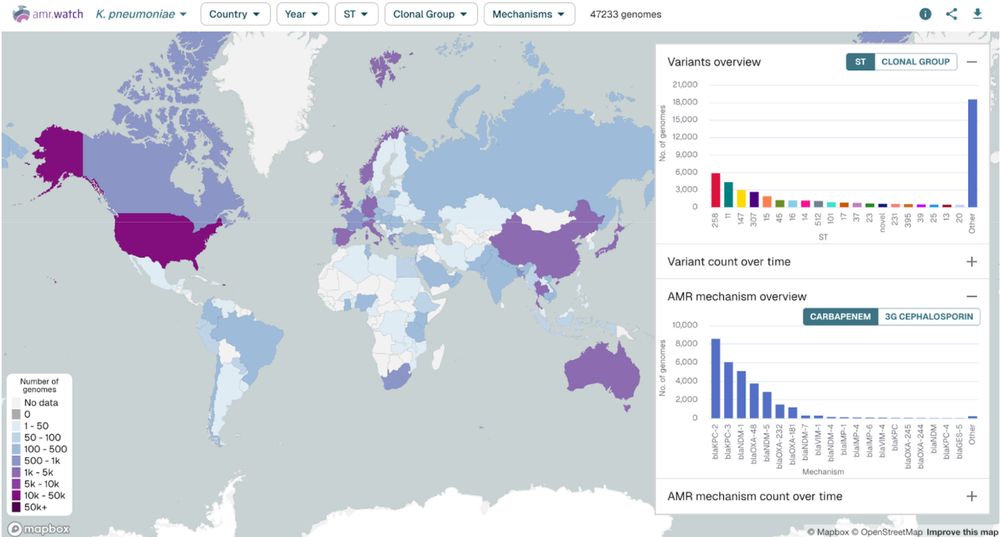
Our latest preprint is out describing amr.watch - a platform for monitoring AMR trends from global genomics data: www.biorxiv.org/content/10.1... (1/6)
22.04.2025 09:54 — 👍 32 🔁 22 💬 1 📌 4
Volume 2 of the #AMR Starter Pack is here!
go.bsky.app/EUY2FUJ
Follow these amazing people along with those in Vol. 1
go.bsky.app/KkZfvrh
And don't forget to pin the Antimicrobial Resistance feed to get all the AMR posts in one place
bsky.app/profile/did:...
02.12.2024 06:52 — 👍 36 🔁 22 💬 8 📌 7
Home
A tool for generating consensus long-read assemblies for bacterial genomes - rrwick/Autocycler
New year, new assemblies!
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
31.12.2024 23:43 — 👍 156 🔁 97 💬 2 📌 3
🚨 New preprint!! How can antibiotic-resistant bacteria increase in abundance in the human gut microbiome, even without antibiotics? 🧐 We show that strain-specific ecological interactions are key to understanding resistance dynamics in microbial communities. 🧵👇 1/5
www.biorxiv.org/content/10.1...
11.12.2024 12:30 — 👍 62 🔁 37 💬 2 📌 5
For more information check out the paper.
This was a great team effort led by Fabrice Graf from LSTM and it's great to see it finally published!
06.12.2024 14:55 — 👍 1 🔁 0 💬 0 📌 0
Together this shows:
(i) that antibiotic susceptibility can re-emerge after an antibiotic stops being used
(ii) that genetic context should be included in AMR databases
(iii) that chloramphenicol could be reintroduced in certain contexts (e.g critically ill with ESBL-E)
6/7
06.12.2024 14:55 — 👍 5 🔁 1 💬 2 📌 1
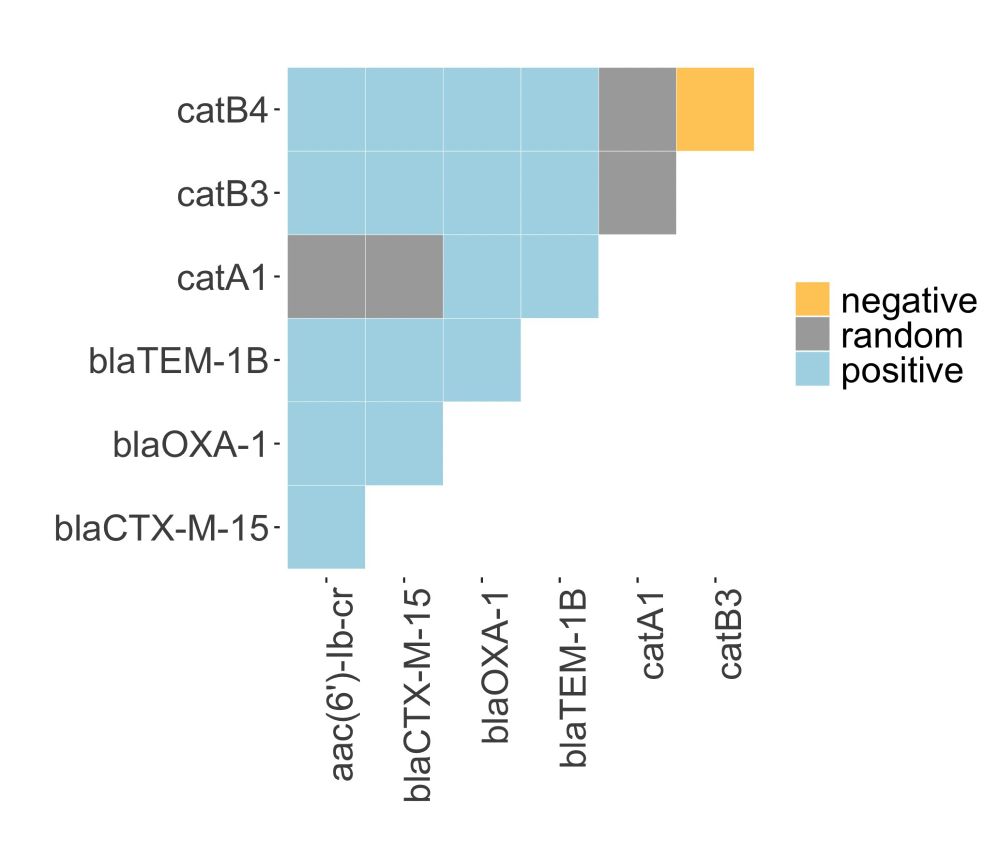
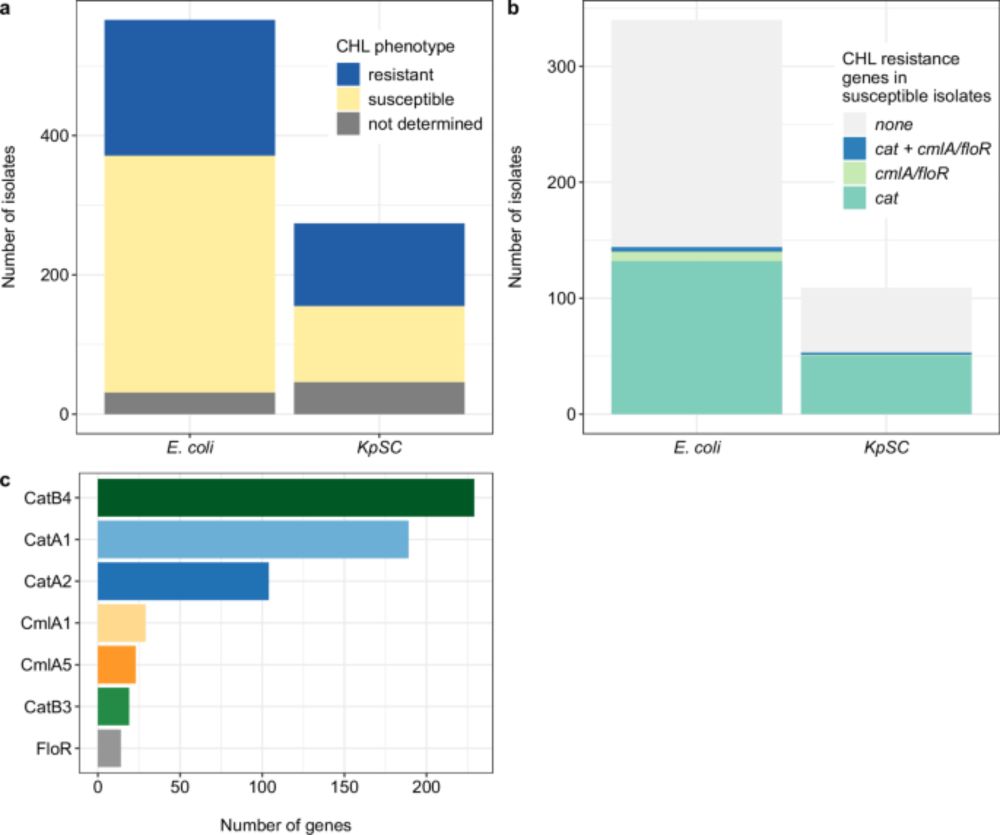
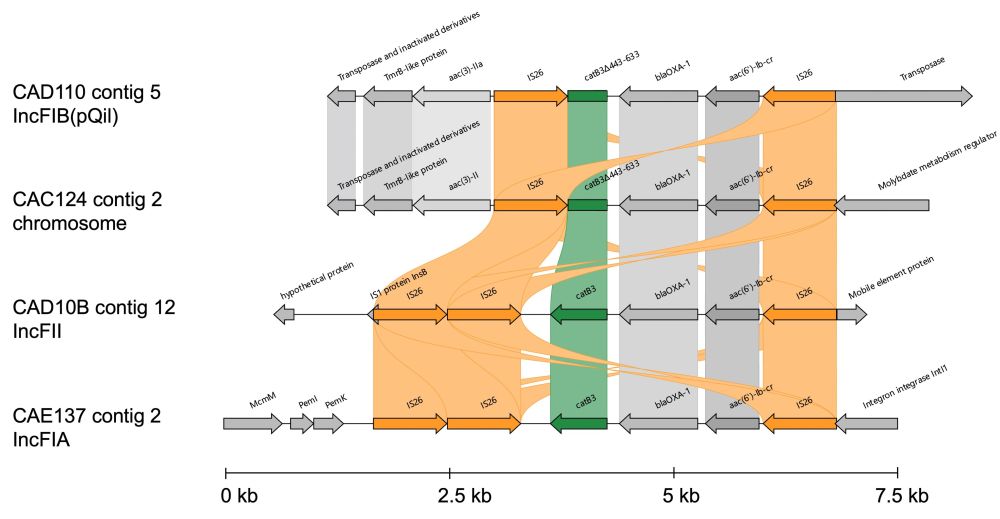
We then used co-occurrence analysis on a larger dataset of short-read E. coli and K. pneumoniae genomes (n=772) which showed that the catB3∆443–633 (i.e. catB4 in figure) and catB3 genes nearly always co-occurred with aac(6')-Ib-cr, blaOXA-1 and the ESBL gene blaCTX-M-15.
5/7
06.12.2024 14:55 — 👍 0 🔁 0 💬 1 📌 0
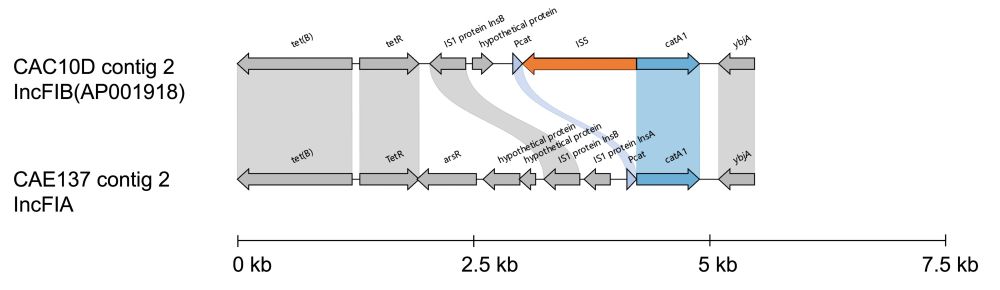
Another IS element, IS5, was found to insert into the promoter region of catA1, again knocking out its function. This shows how it is important to consider the gene environment along with the gene presence or absence in AMR gene detection and surveillance.
4/7
06.12.2024 14:55 — 👍 0 🔁 0 💬 1 📌 0
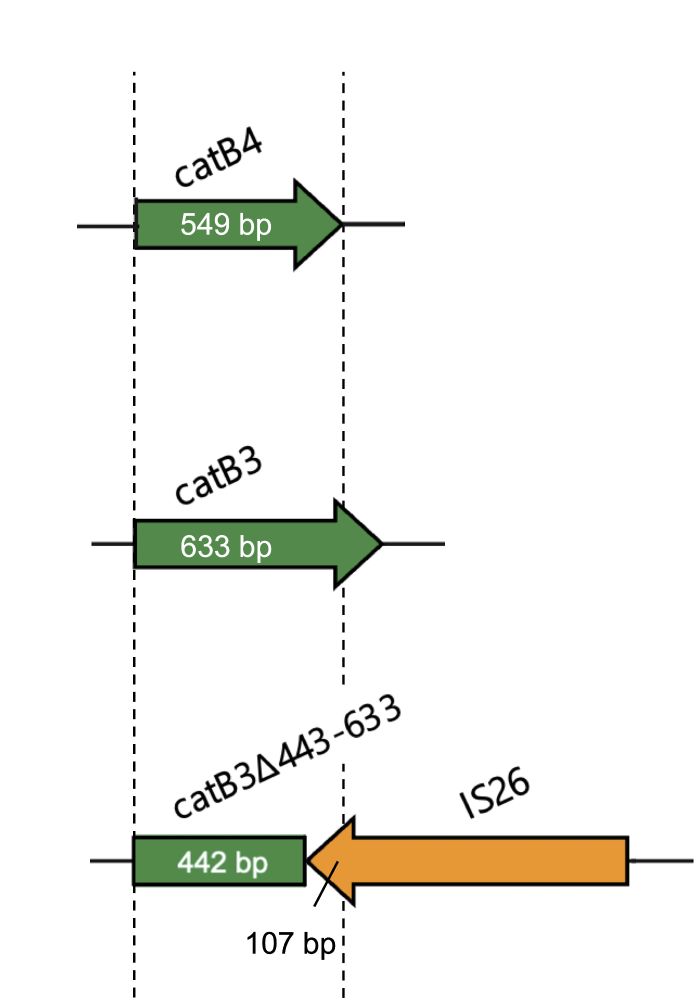
This truncated catB3 gene (catB3∆443–633) appears as catB4 in some AMR gene databases, even though it doesn't confer phenotypic resistance this results in false positives and genotype-phenotype mismatches.
3/7
06.12.2024 14:55 — 👍 0 🔁 0 💬 1 📌 0
Using functional studies and genomics, we found that IS elements were part of the molecular mechanisms driving the rise in chloramphenicol susceptibility. IS26 was found to truncate the chloramphenicol resistance gene catB3, knocking out its function.
2/7
06.12.2024 14:55 — 👍 2 🔁 1 💬 1 📌 0
Senior Editor at Nature Microbiology 💻📄 | handling mycology, parasitology, AMR and phage therapy | https://www.nature.com/nmicrobiol/
The Lancet Microbe is an #OpenAccess journal publishing innovative and practice-changing research and opinion in clinical #microbiology
🌐 thelancet.com/journals/lanmic/home
🧬🦠 Postdoc AMR/Microbial Genomics/Bioinformatics. Tracking AMR bacteria and its MGEs from a One-Health perspective. Passionate ➡️ phylogenomics, GDL, TDL, deep learning, graphs, plasmids, phages,... 🇦🇷 in 🇨🇭
Research Scientist; AI + Biology
thomas-a-neil.github.io
Machine learning and biology. Research Scientist at Google DeepMind. adamgayoso.github.io. Views are my own.
Finished a human genome, working on a few more 👨💻
Lab: https://genomeinformatics.github.io
Posts are my own
DeepMind Professor of AI @Oxford
Scientific Director @Aithyra
Chief Scientist @VantAI
ML Lead @ProjectCETI
geometric deep learning, graph neural networks, generative models, molecular design, proteins, bio AI, 🐎 🎶
Research scientist at @GoogleDeepMind passionate about AI, genomics and biology.
Bioinformatics @UniofAdelaide @BHIresearch
- phages, microbes and more
Texas Medical Center (TMC) Pediatrics Professor-Vaccine Scientist-Author; Member, Philosophical Society of Texas, Texas Academy of Medicine Engineering Science Technology, National Academy of Medicine, American Academy of Arts and Sciences
LSTM takes a whole-picture view on global health issues and works, in partnership, to develop genuine, sustainable, long-term solutions to save and improve lives in the world's most vulnerable communities. Est. 1898.
Antimicrobial resistance and solutions across One Health, professor of microbiology, Maynooth University, Ireland, dog lover.
(she/her) postdoc @ Uni of Exeter in Cornwall 🌊mainly stress out bacteria and track AMR. big fan of the sea, reading, & sunsets 🌞
Researcher in pathogen genomics and evolution
University of Cambridge Vet School @camvetschool.bsky.social
https://pathgenevocam.github.io
Microbiologist, Bacterial evolution, Roslin Institute, University of Edinburgh
Postdoc with Jamie Hall at University of Liverpool, focussing on mobile genetic elements and gene spread in microbial communities! 🦠🧫🧬💻
Prof @Cambridge_Uni Genomic epidemiologist. #Shigella and #AMR enthusiast. Views own. http://gen.cam.ac.uk/baker-group also www.pdu.gen.cam.ac.uk also @pducambridge.bsky.social