
Perspectives in computational mass spectrometry: recent developments and key challenges url: academic.oup.com/bioinformati...
23.01.2026 01:17 — 👍 0 🔁 0 💬 0 📌 0@kozo2.bsky.social
PyData Osaka Organizer, Bioconductor Community Advisory Board, Software Carpentry Japanese Team, 無連想式2ストローク漢字入力方式T-codeユーザー

Perspectives in computational mass spectrometry: recent developments and key challenges url: academic.oup.com/bioinformati...
23.01.2026 01:17 — 👍 0 🔁 0 💬 0 📌 0
Hacking for Metabolomics & Mass Spectrometry format interoperability
www.nfdi4chem.de/metabolomics...

Out now! xcms in Peak Form: Now Anchoring a Complete Metabolomics Data Preprocessing and Analysis Software Ecosystem doi.org/10.1021/acs....
with Phillipine and @jorainer.bsky.social (EURAC), @metabomichael.bsky.social, Hendrik and Norman from @ipbhalle.bsky.social, @janstanstrup.bsky.social, et al.
Great work from @philouail.bsky.social 🙌
#xcms now fully integrated into @bioconductor.bsky.social 💪
👉 #metabolomics #MassSpectrometry #rstats
 01.12.2025 18:59 — 👍 1 🔁 1 💬 0 📌 0
01.12.2025 18:59 — 👍 1 🔁 1 💬 0 📌 0

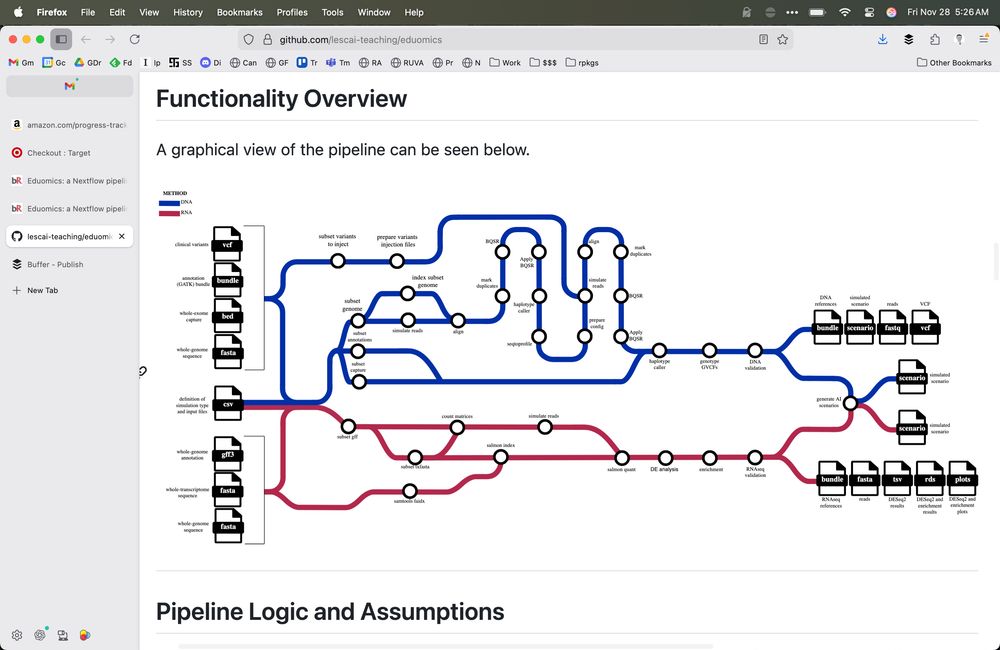
Eduomics: a Nextflow pipeline to simulate -omics data for education www.biorxiv.org/content/10.1... 🧬🖥️🧪 github.com/lescai-teach...
01.12.2025 19:02 — 👍 11 🔁 3 💬 0 📌 1
Upcoming community sessions this week
carpentries.org/community/ev...
Events are in UTC Time :
📣 Workbench Transition Coworking
24 Nov 09:00 & 19:00
📣 👋 Welcome Session with The Carpentries
24 Nov 12:00
🇬🇧 UK Carpentries Community Call
24 Nov 16:00
🕸️ Creating a workshop website
25 Nov 09:00

🌍 We are proud to share highlights from Ethiopia’s first in-person Bioconductor workshop, held in Addis Ababa from 25-29 Aug 2025!
The course united 26 participants for five days of hands-on training in R/Bioconductor and bulk RNASeq analysis.
🔗 Read more: blog.bioconductor.org/posts/2025-1...

Reactome Knowledgebase 2026 url: academic.oup.com/nar/article/...
24.11.2025 13:54 — 👍 1 🔁 0 💬 0 📌 0
Open Enzyme Database: a community-wide repository for sharing enzyme data url: academic.oup.com/nar/article/...
18.11.2025 18:49 — 👍 1 🔁 0 💬 0 📌 0
MassBank: an open and FAIR mass spectral data resource url: academic.oup.com/nar/article/...
16.11.2025 13:19 — 👍 1 🔁 0 💬 0 📌 0
DancePartner: Python Package to Mine Multiomics Relationship Networks from Literature and Databases | Journal of Proteome Research pubs.acs.org/doi/abs/10.1...
08.11.2025 13:37 — 👍 1 🔁 0 💬 0 📌 0
MSOne: An AI-powered software suite enabling end-to-end analysis of high-resolution LCMS data for metabolomics data mining | ChemRxiv - doi.org/10.26434/che...
03.11.2025 17:04 — 👍 1 🔁 0 💬 0 📌 0
Practical Guidance for Training Machine Learning Models in Metabolomics and Mass Spectrometry Research | Analytical Chemistry pubs.acs.org/doi/full/10....
26.10.2025 10:59 — 👍 3 🔁 2 💬 0 📌 0
RDMkit: A research data management toolkit for life sciences: Patterns www.cell.com/patterns/ful...
21.10.2025 13:19 — 👍 1 🔁 0 💬 0 📌 0
From @plos.org #Computational #Biology | Ten quick tips for developing a reproducible #Shiny application | #Bioinformatics #Education #PLOSCBQT #OpenScience #OpenSource 🧬 🖥️ 🧪🔓
⬇️
journals.plos.org/ploscompbiol...
📣 HPC Carpentry Community Call
16 Oct 11:00 & 21:00.

🛡️ Applications are now open to join the Bioconductor Code of Conduct Committee 2025!
Be part of a thoughtful, community-driven team that meets 3–5 times a year to ensure Bioconductor remains a respectful, inclusive, and thriving space 🌱
👉 Apply by October 31, 2025:
docs.google.com/forms/d/e/1F...
Did you miss this.... Introducing Partial Spectra DB 🧪—an open-access database for partially annotated lipids!
✨ Explore curated spectra - Browse, Search, and Download data for your workflows.
🌍 Got spectra? Contribute & help grow this community-driven resource!
🔗 www.lipidmaps.org/databases/pa...

Checkout this article I found at PLOS: dx.plos.org/10.1371/jour...
12.10.2025 11:12 — 👍 2 🔁 0 💬 0 📌 0
Thank you for citing #tidyplots 🙏
Jakub Idkowiak et al. Best practices and tools in R and Python for statistical processing and visualization of lipidomics and metabolomics data. Nature Communications (2025).
doi.org/10.1038/s414...
#rstats #dataviz #phd

mzPeak: Designing a Scalable, Interoperable, and Future-Ready Mass Spectrometry Data Format | Journal of Proteome Research pubs.acs.org/doi/full/10....
05.10.2025 16:58 — 👍 1 🔁 0 💬 0 📌 0Release Notes for Cytoscape 3.10.4
cytoscape.org/release_note...

Supervised Contrastive Learning Leads to More Reasonable Spectral Embeddings | Analytical Chemistry pubs.acs.org/doi/10.1021/...
19.09.2025 03:22 — 👍 2 🔁 0 💬 0 📌 0
Intelligent Tool Orchestration for Rapid Mechanistic Model Prototyping: MCP Servers as AI-Biology Interfaces
www.biorxiv.org/content/10.1...

Koza and Koza-Hub for born-interoperable knowledge graph generation using KGX
arxiv.org/abs/2509.09096
Interoperability of Bioconductor packages within the ELIXIR Research Software Ecosystem using the EDAM ontology - CNRS - Centre national de la recherche scientifique cnrs.hal.science/hal-05250261v1
16.09.2025 05:19 — 👍 2 🔁 0 💬 0 📌 0
SBMLtoOdin and menelmacar: Interactive visualisation of systems biology models for expert and non-expert audiences url: academic.oup.com/bioinformati...
08.09.2025 13:14 — 👍 0 🔁 0 💬 0 📌 0
Understanding Data Analysis Steps in Mass-Spectrometry-Based Proteomics Is Key to Transparent Reporting | Journal of Proteome Research pubs.acs.org/doi/full/10....
08.09.2025 10:07 — 👍 0 🔁 0 💬 0 📌 0
Reverse Spectral Search Reimagined: A Simple but Overlooked Solution for Chimeric Spectral Annotation | Analytical Chemistry pubs.acs.org/doi/abs/10.1...
19.08.2025 12:44 — 👍 0 🔁 0 💬 0 📌 0