AlphaFold’s success builds on decades of tech advances, massive efforts to collect, curate, and share structural data, and rigorous benchmarking. What will it take to see a similar leap in structural ensemble prediction? @stephanieaw.bsky.social and I share some thoughts here 👇 Feedback is welcome!
07.05.2025 04:49 — 👍 21 🔁 5 💬 1 📌 0
Anyhow, thanks for your efforts.
24.03.2025 14:11 — 👍 0 🔁 0 💬 0 📌 0
That would be awesome.
24.03.2025 14:11 — 👍 0 🔁 0 💬 0 📌 0
However, do you think it would be better to do 5000 sample compared to 1000 sample?
And what do you think would be faster to run it on a local machine with RTX 4090 or keep on the colab with the A100?
21.03.2025 22:01 — 👍 0 🔁 0 💬 1 📌 0
Fourth, facilitating the exploration of potential conformations before deciding which to simulate further so instead of simulations that takes weeks you will do the modelling of Bioemu than decide what to do next,
Fifth, it can compare the wild versus mutant sequence configurations
21.03.2025 21:59 — 👍 1 🔁 0 💬 1 📌 0
Second it can help you to scan the preferred conformation spaces of protein sequence to show you if there is a preferred configuration or multiple configurations,
Third it can shows the most unstable part of protein so you can investigate there if needed,
21.03.2025 21:58 — 👍 1 🔁 0 💬 1 📌 0
Hello Pablo,
First I need to thank and your team for this cool technology
And I like to summarise the uses I can see
it’s a great tool to check the stability of your structure (protein or peptide) as a first point,
21.03.2025 21:57 — 👍 2 🔁 0 💬 1 📌 0
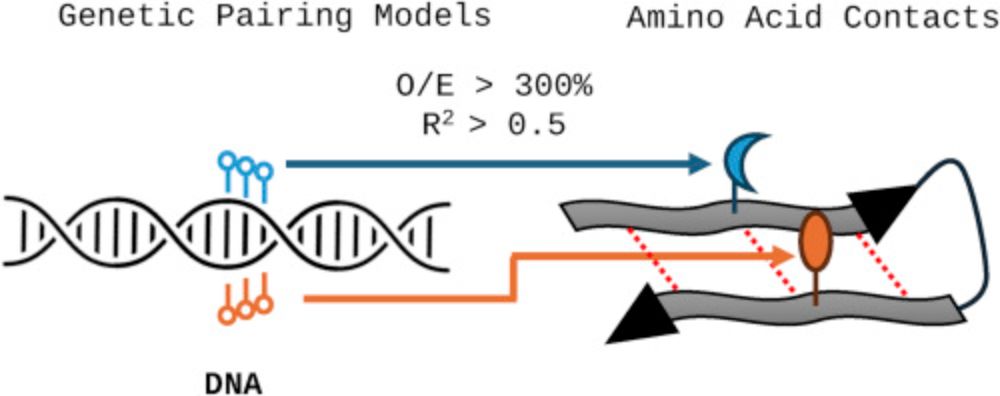
where we prepose a "proteomic code" derived from DNA's genetic code, suggesting that specific amino acid pairings determine protein folding (intra-connections) and protein-protein interactions (inter-connections) based on DNA and biophysics.
19.03.2025 15:59 — 👍 0 🔁 0 💬 0 📌 0
Prof @sydneyuni.bsky.social Engineer and scientist building bridges between research & industry at USYD & @ansto.bsky.social, Australia.
She/Her. Mum & wife. Leader & Mentor. Loving ⛷🏄♀️ & life in general! Comments are my own
Postdoc @ ETH Zurich / SIB Swiss Institute of Bioinformatics | Protein bioinformatics
Theoretical physicist interested in the physics of living systems and statistical physics. Professor at LMU Munich. Passionate about emergent phenomena and interdisciplinary research.
https://www.theorie.physik.uni-muenchen.de/lsfrey/
Postdoc - Boston University | FeS clusters, EPR and redox chemistry 🧪
Research Fellow @flatironinstitute.org @simonsfoundation.org
Formerly @csail.mit.edu @msftresearch.bsky.social @uconn.bsky.social
Computational systems x structure biology | he/him | https://samsl.io | 👨🏼💻
💻 We are the Structural Bioinformatics and Computational Biology track of the International Society for Computational Biology (ISCB)
InterPro provides functional analysis of proteins by classifying them into families and predicting domains and important sites.
ebi.ac.uk/interpro/
#cryoem #teamtomo #volumeEM #thermofisherscientific
Yosemite is my happy place
Philosopher of science and Co-Director of the Munich Center for Mathematical Philosophy at LMU Munich. Interested in formal epistemology, social epistemology, philosophy of physics and cognitive science. Website: http://stephanhartmann.org
PhD student at @biozentrum.bsky.social. I prefer protein interactions to human ones.
We aim at unraveling the intricacies of molecular perception and designing sensing molecular systems from the ground up. We work at the interface of biophysics, biochemistry, and materials science.
@ Max Planck Institute for Polymer Research, Mainz.
Math Assoc. Prof. at Aix-Marseille (France)
Currently on Sabbatical at CRM-CNRS, Université de Montréal
https://sites.google.com/view/sebastien-darses/welcome
Teaching Project (non-profit): https://highcolle.com/
#compchem Research Scientist at Vita Nova Institute- Work interests mostly related to the simulation of biomolecular systems - Lead developer of the Making It Rain, Cloud-Bind and ParametrizANI projects.
https://pablo-arantes.github.io/
UW biology prof.
I study how information flows in biology, science, and society.
Book: *Calling Bullshit*, http://tinyurl.com/fdcuvd7b
LLM course: https://thebullshitmachines.com
Corvids: https://tinyurl.com/mr2n5ymk
I don't like fascists.
he/him
Structural Biologist | Accelerating drug discovery
Obsessed with X ray-Crystallography and Cryo-EM
Sharing: Practical Structural Bio Tips
Account of the Max Planck Institute for the Physics of Complex Systems in Dresden; tweets by Pablo Pérez, Uta Gneisse, and Pierre Haas @lepuslapis.bsky.social.
UCSF ChimeraX - software for visualizing biomolecules
Assistant Professor at UMich with a focus on computational pharmacology (he/him)
Our research group at the University of Ottawa specializes in protein engineering and computational protein design.
mysite.science.uottawa.ca/rchica/
protein geek, founder and CEO of AI Proteins @aiproteins.com
https://linkedin.com/in/cdbahl