This paper was a truly @uchichemistry.bsky.social team effort, jointly led by Shannon Lu, Matt Styles, and Frank Gao with me, Aaron Dinner, and @vaikuntsuri.bsky.social – along with a critical support cast. 12/12
15.10.2025 17:19 — 👍 3 🔁 0 💬 0 📌 0
On the flip side, proteins can evolve to be “hard” to bind – thereby tuning their inherent propensity for forming new interactions! 11/12
15.10.2025 17:19 — 👍 3 🔁 0 💬 1 📌 0
From an evolutionary perspective, this work uncovers that the likelihood of new interactions evolving with a target protein is largely dictated by the protein itself. Meaning proteins can evolve to be “bindable” – and poised for the evolution of new interactions. 10/12
15.10.2025 17:19 — 👍 2 🔁 0 💬 1 📌 0

Why does this matter? We present a conceptual, experimental, and computational framework to understand selection success in binder discovery. We turn what is usually more of an art into a science. 9/12
15.10.2025 17:19 — 👍 3 🔁 0 💬 2 📌 0

Machine learning trained on our data – critically using contrastive learning with our vast non-binder datasets - can successfully predict binders, but needs >50% of the data to work well - showing you need large datasets to understand these vast and diverse landscapes. 8/12
15.10.2025 17:19 — 👍 2 🔁 0 💬 1 📌 0

Each target has a unique "fitness landscape" with distinct binding clusters. These landscapes are highly reproducible - run the same selection twice, get the same answer in terms of fitness landscapes and clusters – even though the specific sequences will be different every time! 7/12
15.10.2025 17:19 — 👍 1 🔁 0 💬 1 📌 0

The simplest randomization strategy (using all 20 amino acids equally) is nearly optimal! Fancy biased designs showed negligible improvements. Nature already optimized this. 6/12
15.10.2025 17:19 — 👍 2 🔁 0 💬 1 📌 0

We found target identity matters WAY more than how you design your library. "Easy" targets need only 2-3 amino acids; "hard" targets need 6+ fixed position or have strict topographical requirements. 5/12
15.10.2025 17:19 — 👍 2 🔁 0 💬 1 📌 0

Why are some targets “easy” and some “hard”? Minimal binding motifs of just 2-8 specified amino acids determine success. Fewer amino acids needed in a motif = a more common motif in a theoretical library = easier to find binders. It's surprisingly simple! 4/12
15.10.2025 17:19 — 👍 2 🔁 0 💬 1 📌 0

We created a framework to quantify “target difficulty” and found the likelihood of a random protein variant binding a target varies WILDLY - from 1 in 10,000 to over 1 in 10 billion! Most targets have a binding frequency around 1 in 100 million. 3/12
15.10.2025 17:19 — 👍 2 🔁 1 💬 1 📌 0

To answer this, we combined our PANCS-Binder tech with advanced computational approaches to perform and analyze >1,300 selections testing how random protein variants bind to 96 different targets. 2/12
15.10.2025 17:19 — 👍 2 🔁 0 💬 1 📌 0


Thank you so much to the University of Kansas Department of Chemistry, Chembio program, COBRE program, and most importantly the trainees who invited me to take part in the 4th Annual Chemical Biology Symposium. Fantastic science and a wonderful way to spend a weekend. And congrats to awardees!!
11.10.2025 22:12 — 👍 5 🔁 2 💬 0 📌 0

They've done studies, you know.
60% of the time it works every time.
Good balanced take on AI in protein design...hype and potential:
blog.genesmindsmachines.com/p/we-still-c...
07.10.2025 16:36 — 👍 4 🔁 0 💬 0 📌 0

A Warm Welcome to Our Newest UChicago Chemists! As an incoming student, you're now at the epicenter of chemical innovation. We're excited to have you become part of our story and see the discoveries you will make this year.
29.09.2025 18:07 — 👍 6 🔁 2 💬 0 📌 0
Excited to hear about the amazing research from the @chembiobryan.bsky.social group on taming acyl chlorides for #ChemBio applications from the first author of the @natchem.nature.com paper, @shubha-pani.bsky.social.
Register for free for the 5th Virtual #ChemBioTalks to join: cvent.me/G1geWW
29.09.2025 04:57 — 👍 9 🔁 2 💬 0 📌 0

The third and final Flash Talk of the 5th Virtual #ChemBioTalks will be given by Shubhashree Pani (@shubha-pani.bsky.social) from the group of @chembiobryan.bsky.social: "Masked acylating agents for proximity labeling of biomolecules" www.nature.com/articles/s41...
#ChemBio #ChemSky #DrugDiscovery
29.09.2025 04:49 — 👍 7 🔁 2 💬 0 📌 2
Science in the US is primarily funded directly through the departments (NIH, NSF, DOD, etc), philanthropy, and industry. Not sure what you are asking?
23.09.2025 15:17 — 👍 0 🔁 0 💬 1 📌 0
Sure. Private sector also funds science! More science is great!
23.09.2025 14:48 — 👍 0 🔁 0 💬 1 📌 0
Thank you! The main message is aside from the direct health and tech benefits, every dollar invested in science brings ~$2.50 back to the country (across every state!). As far as tax dollars go, that is a great investment. Rest assured we (scientists) take this public trust very seriously.
23.09.2025 14:34 — 👍 0 🔁 0 💬 1 📌 0
Nope! For sure not. See above thread.
23.09.2025 14:24 — 👍 0 🔁 0 💬 1 📌 0

The NIH is a sound investment for the US taxpayer
Research funded by the National Institutes of Health is essential for improving the health of Americans and developing new drugs and treatments for a wide range of diseases. Research organism: None
You cannot use grants to pay for dinners! The grant pays for science. Here is some good reading: pmc.ncbi.nlm.nih.gov/articles/PMC... The picture shows a celebration of a group of scientists excited about working hard so our country continues to lead in innovation. That’s good for everyone.
23.09.2025 14:23 — 👍 0 🔁 0 💬 0 📌 0
What are you talking about? That’s not how anything works…
23.09.2025 14:10 — 👍 0 🔁 0 💬 2 📌 0

A successful NSF grant in 2025 deserves a nice dinner celebration. Thanks to this incredible crew of rockstars
23.09.2025 01:10 — 👍 51 🔁 1 💬 1 📌 0

Discovery of Macrocyclic Peptide Binders, Covalent Modifiers, and Degraders of a Structured RNA by mRNA Display
RNA targeting represents a compelling strategy for addressing challenging therapeutic targets that are otherwise intractable through traditional protein targeting. Revolutionary approaches in RNA-focused small molecule libraries have successfully identified RNA-binding ligands but generally remain limited in diversity and impeded by a dearth of structural insight into RNA and RNA complexes. Cyclic peptides are potential structural mimics of evolutionary RNA-protein interacting motifs and can be massively diversified and selected via genetically encoded libraries, offering a complementary approach. This study introduces genetically encoded thioether cyclic peptide libraries constructed through mRNA display using a dibromoxylene linker and its fluorosulfonyl derivative that can covalently engage RNA nucleophiles. Using an optimized mRNA display workflow for RNA binders, we discovered high affinity, covalent and noncovalent binders for SNCA 5′ UTR IRE, the upstream iron-responsive element that post-transcriptionally regulates the expression of α-synuclein, an intrinsically disordered protein implicated in Parkinsonism and related neurodegenerative diseases. Notably, a stringent selection strategy employing “base-paired” target analog counterselection enhanced specificity by deenriching nonspecific electrostatic interactions mediated by polycationic residues. Further engineering hit peptides with an imidazole tag yielded selective RNA degraders in which covalent degraders showed noticeably improved potency from noncovalent counterparts. This work provides a prototype framework for evolution-driven, high-throughput, RNA-targeted drug discovery using cyclic peptides.
Excited to share our most recent work out in @jacs.acspublications.org today! We combined mRNA display with macrocyclic peptide chemistry to discover novel RNA-targeting molecules. This fits into our mission to target RNA regulation with novel therapeutic modalities
pubs.acs.org/doi/10.1021/...
15.09.2025 14:47 — 👍 30 🔁 5 💬 0 📌 0
Update from Chicago. The situation is dire. Complete anarchy. Please send the national guard asap to put an end to this lawlessness
31.08.2025 20:42 — 👍 26 🔁 4 💬 3 📌 0
So excited to have this great collaborative work out!!! It's been a delight working with the @jeffmartell.bsky.social group. More to come for sure!
28.08.2025 19:51 — 👍 6 🔁 2 💬 0 📌 0
Congrats to my amazing colleagues John and Dmitri!!! What an inorganic/materials duo powerhouse 💪💪💪.
20.08.2025 22:54 — 👍 0 🔁 0 💬 0 📌 0
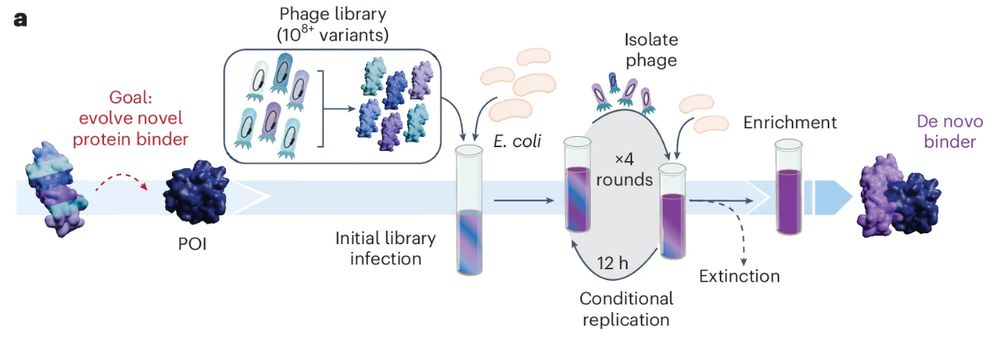
PANCS-Binders (phage-assisted noncontinuous selection of protein binders) screens multiple high-diversity protein libraries against a panel of dozens of targets for high-throughput binder discovery. @chembiobryan.bsky.social @mstyles-chembiol.bsky.social
www.nature.com/articles/s41...
06.08.2025 19:49 — 👍 24 🔁 14 💬 1 📌 2
I run, ski, and engineer cells and materials; asking how nutrient sensing & signaling work: bit.ly/RK-GoogleScholar
🇪🇪: TalTech| Ex: Tartu Ülikool; 🇸🇪: Karolinska Institutet, Chalmers; 🇯🇵: Kyutech; 🇮🇳: IITD
We use structure-guided engineering to design biologics & immunotherapies.
Associate Professor of Immunology,
Director - Center for Antibody & Protein Engineering (CAPE) @ Moffitt Cancer Center
www.thelucalab.org
Official account of the College of Chemistry at the University of California, Berkeley🔬📚
Chemist, Dad, Group Leader, Associate Professor. Excited about novel chemical modalities in drug discovery
MSc student in Bioinformatics
Biomedical Science and music degrees.
Professional interests: Molecular dynamics, Bioinformatics, Medical Microbiology, Ludomusicology
Music, Guitar, Gaming, LARP, World of Warcraft, BJJ, Judo 🏳️🌈🏳️⚧️
he/they
Editor of Tetrahedron Chem
Chemical biology, CRISPR and beta-cell biology @ Broad Institute | Harvard Medical School | Brigham and Women's Hospital
Assistant Professor, UW-Madison Chemistry. Group website: http://martellgroup.chem.wisc.edu.
Mobilizing the fight for science and democracy, because Science is for everyone 🧪🌎
The hub for science activism!
Learn more ⬇️
http://linktr.ee/standupforscience
Drugging intrinsically disordered proteins
CEO/CSO/Co-Founder, Bind Research
PI, eXtreme Dynamics & Therapeutics Lab, UCL
(she/her)
Chemical biologist broadly defined
A chembio group @Temple Chemistry & Fox Chase Cancer Center. We work on small molecules, peptides & protein probes, expanding toolkits for proteomics & cellular imaging, with the goal of deciphering cell biology. Find more @ https://rosswang.weebly.com
Professor @ Scripps Research | Department of Chemistry | chemical biology & chemical proteomics
Head of the Budin lab at UCSD (www.budinlab.com). Musings on thin layers of grease in our cells (and other topics). Lipids, cell membranes, biophysics, chem bio, evolution. 🏳️🌈
Student-run account of the Michelle Chang Lab at Princeton University || Enzymology || Metabolic Engineering || Biocatalysis || Synthetic Biology
Assistant Professor of Molecular Biology, Center for Alzheimer's and Neurodegenerative Diseases, UT Southwestern. My lab studies damaged things. Sometimes we fix them. Hiring! Write if interested in postdoc or tech position. https://www.davidwsanders.com
News and updates from the Sletten Group at @UCLA @uclachem | student-run account
https://slettengroup.chem.ucla.edu
Associate Dean for data science, and Professor of Medicine and Human Genetics at the University of Chicago.


















