

Such a treat to host @gioelelamanno.bsky.social (EPFL) today and be the first to hear more about his lab's latest work, posted just two days ago on bioRxiv.
Missed the talk?
Here's the preprint: https://www.biorxiv.org/content/10.1101/2025.10.13.682018v1
16.10.2025 17:52 — 👍 6 🔁 2 💬 1 📌 0

ikea-style logo of splongget
1/ First preprint from @jdemeul.bsky.social lab 🥳! We present our new multi-modal single-cell long-read method SPLONGGET (Single-cell Profiling of LONG-read Genome, Epigenome, and Transcriptome)! www.biorxiv.org/content/10.1...
10.09.2025 15:48 — 👍 47 🔁 16 💬 1 📌 1
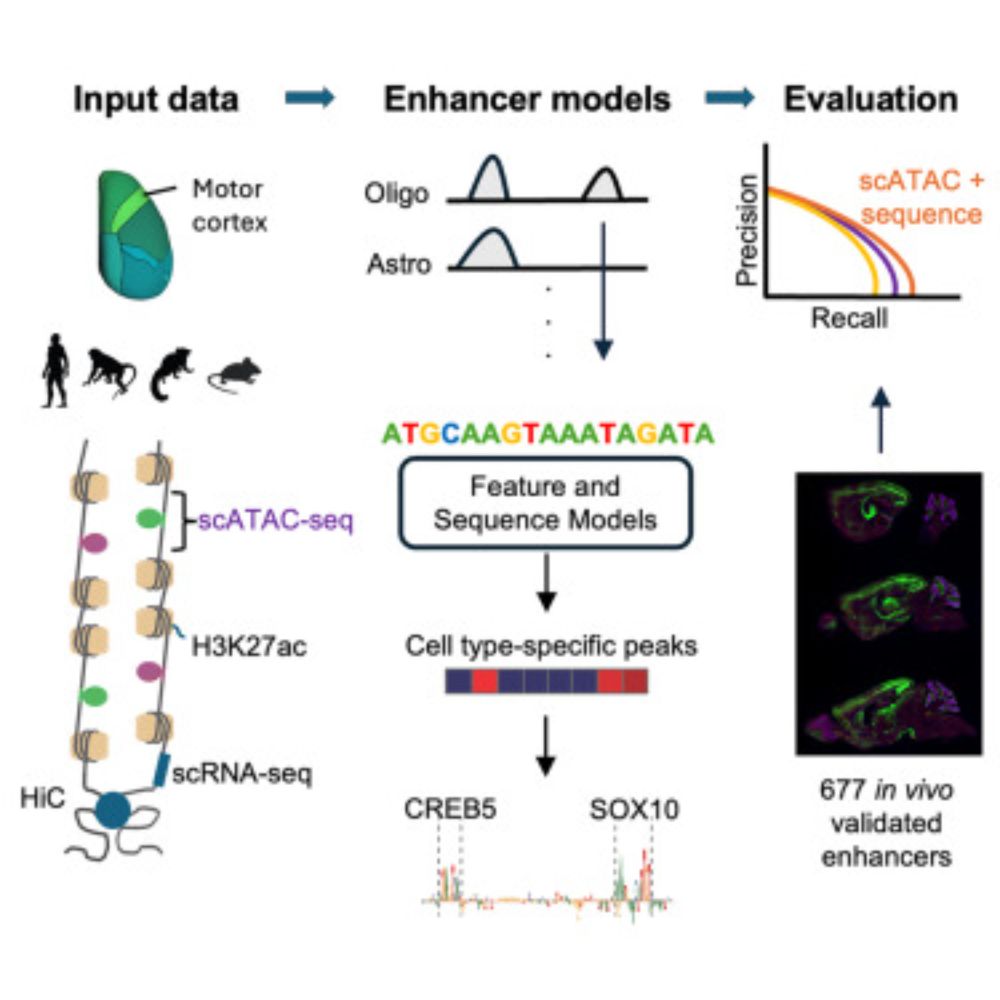
Evaluating methods for the prediction of cell-type-specific enhancers in the mammalian cortex
Johansen et al. report the results of a community challenge to predict functional
enhancers targeting specific brain cell types. By comparing multi-omics machine learning
approaches using in vivo data...
Check out our work on evaluating methods for predicting in vivo cell enhancer activity in the mouse cortex! Combined, scATAC peak specificity and sequence-based CREsted predictions gave the best predictive performance, aiming to advance genetic tool design for cell targeting in the brain.
21.05.2025 16:45 — 👍 20 🔁 10 💬 1 📌 0

VIB.AI's International PhD call is now open!
https://tinyurl.com/53nc9vnd
If you're interested in applying AI to biology, take a look at the PhD projects across our labs.
🗓️ Deadline: June 22nd
06.05.2025 14:48 — 👍 6 🔁 7 💬 0 📌 0

We released our preprint on the CREsted package. CREsted allows for complete modeling of cell type-specific enhancer codes from scATAC-seq data. We demonstrate CREsted’s robust functionality in various species and tissues, and in vivo validate our findings: www.biorxiv.org/content/10.1...
03.04.2025 14:30 — 👍 75 🔁 38 💬 1 📌 5
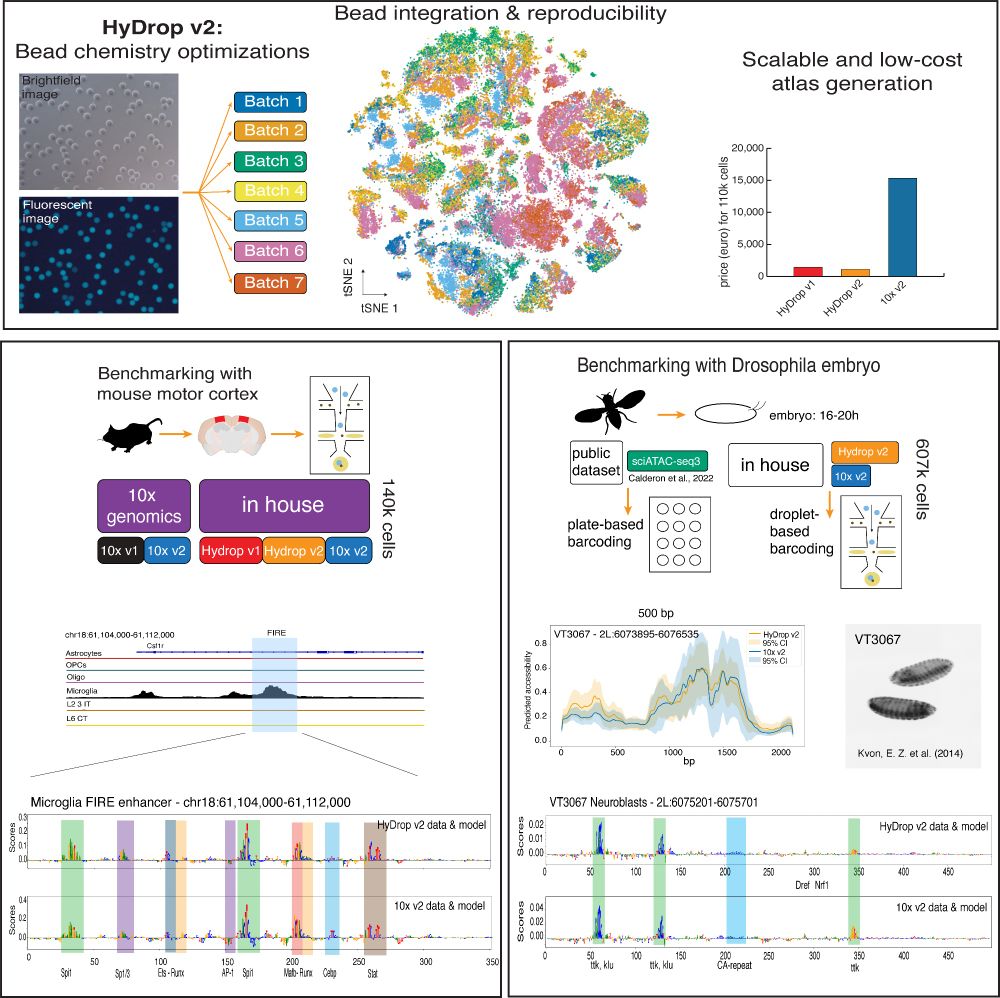
Data collected with the new sequencing platform HyDrop v2 is shown. First, a schematic overview of the bead batches of the microfluidic beads is followed by a tSNE and a barplot showing the costs in comparison to 10x Genomics.
Then, a track of mouse data (cortex) is shown together with nucleotide contribution scores in the FIRE enhancer in microglia. Here, the HyDrop and 10x based models show the same contributions.
On the right, the Drosophila embryo collection is explained; in the paper HyDrop v2 and 10x data are compared to sciATAC data. Then, a nucleotide contribution score is also shown, whereas HyDrop v2 and 10x models show the same contribution, just as in mouse.
Our new preprint is out! We optimized our open-source platform, HyDrop (v2), for scATAC sequencing and generated new atlases for the mouse cortex and Drosophila embryo with 607k cells. Now, we can train sequence-to-function models on data generated with HyDrop v2!
www.biorxiv.org/content/10.1...
04.04.2025 08:52 — 👍 55 🔁 25 💬 2 📌 2
Isn't Nestle Swiss? Why boycott this brand specificaly?
04.03.2025 16:33 — 👍 0 🔁 0 💬 1 📌 0

Ziga Avsec (Google DeepMind), Julien Gagneur (TU Munich), Tanja Kortemme (UCSF), David Kelley (Calico), Ben Lehner (Sanger), Franca Fraternali (UCL), Oliver Stegle (EMBL/DKFZ), and Amos Tanay (Weizmann)
Looking forward to the Inaugural Symposium of the Center for AI & Computational Biology vib.ai with a great line-up of speakers at the interface of AI & biology: D. Kelley, J. Gagneur, Z. Avsec, T. Kortemme, B. Lehner, F. Fraternali, A. Tanay & O. Stegle (20Nov) www.vibconferences.be/events/vibai...
12.11.2024 11:26 — 👍 38 🔁 17 💬 1 📌 0
Professor at #EPFL - #RNAVelocity inventor - Laboratory of Brain Development and Biological Data Science. Single-cell and spatial biologist studying brain development and lipids. #ERCStg investigator
fly person, PhD student @ Stein Aerts Lab of Computational Biology, VIB.AI
Postdoc at @CBD KU Leuven 🇧🇪. Interested in translation, structural biology, ageing and everything involving proteins
🔬 Spatial Catalyst @ VIB Technologies
📍Ghent
Empowering researchers with expert spatial omics solutions – transcriptomics, proteomics & beyond.
My favorite ice cream flavors are science & feminism. PhD candidate at VIB.AI in Stein Aerts lab 🪰 - she/her
EMBO postdoc @teichlab @SCICambridge | Single-cell genomics and gene regulation | PhD in evodevo genomics @kaessmannlab @UniHeidelberg
VIB offers top-tier training and conference programs designed to inspire creativity, collaboration, and lifelong learning, enhancing research excellence and career development for scientists globally. Find out more: vibtrainingandconferences.be
Author: Children of Time, Shadows of the Apt, Final Architecture, City of Last Chances and others. adriantchaikovsky.com
Subterranean Press creates readable art, publishing luxurious specialty, limited editions and groundbreaking original works in the science fiction, fantasy, and horror genres. Dispatches from the warehouse and other news. http://linktr.ee/subterraneanpress
PhD student
Laboratory of computational biology
VIB.AI & VIB-KU Leuven center for Brain & Disease research 🧬🧬🧬
Proteomics Expertise Unit Leader at VIB-KU Leuven Center for Brain & Disease Research
Proteomics Expert at KU Leuven Laboratory of Applied Mass Spectrometry
PhD Student at Stein Aerts Lab of Computational Biology.
Stem cell and developmental biologist, Professor @ University of Leuven, Belgium. #Pluripotency #Epigenetics
Postdoc EMBO fellow @Treutlein lab ETH Zurich
Previously @Aerts lab VIB-KU Leuven
Interested in Gene regulation + Evolution, Fruit fly enthusiast
PhD student @ Stein Aerts Lab of Computational Biology.
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Assistant Professor UMass Chan
Previously IMP Vienna, Stanford Genetics, UW CSE.
Researcher at FU Berlin •Nature's 10 2024 •Creator of RW #Hijackedjournals checker •Scientific misconduct •Plagiarism • #papermills •Corruption in higher education
Author (The Fault in Our Stars, The Anthropocene Reviewed, etc.)
YouTuber (vlogbrothers, Crash Course, etc.)
Football Fan (co-owner of AFC Wimbledon, longtime Liverpool fan)
Opposed to Tuberculosis








