Papers are like buses... You wait for ages, then two come along at once.
Huge congrats to @bornanovak.bsky.social and @jefflotthammer.bsky.social for pushing and driving every aspect of this work, preprinted ~1 year ago to the day (Friday before BPS), now published!
www.nature.com/articles/s41...
19.02.2026 03:30 —
👍 86
🔁 35
💬 6
📌 2
1/ Excited to share a new preprint from the lab! We identify NSD3 as a novel regulator of chromatin 3D structure. 🧵
11.02.2026 03:21 —
👍 12
🔁 4
💬 2
📌 0
Pleased to share the final version of this behemoth of a paper, now finally published. I guess I can retire now?
www.nature.com/articles/s41...
More functional data, many thousands of words removed, and a few other updates from last year's preprint.
12.02.2026 11:22 —
👍 134
🔁 57
💬 7
📌 5

Lab lunch at Truth BBQ! A Houston classic!!
14.11.2025 18:09 —
👍 2
🔁 0
💬 0
📌 0
Congratulations Bruno, Patri, Justin and teams!!!!
05.11.2025 01:21 —
👍 1
🔁 0
💬 0
📌 0

Selective RNA sequestration in biomolecular condensates directs cell fate transitions - Nature Biotechnology
Stem cell differentiation is controlled by manipulating RNA condensates.
1/ Excited to share our new study with @brumbaugh-lab.bsky.social, out in @natbiotech.nature.com! P-bodies selectively sequester RNAs encoding cell fate regulators, often from the preceding developmental stage. Releasing these RNAs can drive changes in cell identity. 🧵 www.nature.com/articles/s41...
28.10.2025 16:02 —
👍 93
🔁 37
💬 4
📌 6
Congratulations Josh and team!!!!!!
05.11.2025 01:19 —
👍 1
🔁 0
💬 1
📌 0

(1/10) How do diverse leukemia mutations converge on the same molecular program? In #RibackLab first manuscript @cp-cell.bsky.social, collaboration with @goodell-lab.bsky.social shows that disparate mutations rewire shared protein networks to form nuclear condensates called C-bodies.
04.11.2025 17:58 —
👍 51
🔁 23
💬 7
📌 4
Happy to share the latest work from the lab, led by @mudgal17.bsky.social, in collaboration with the Weis lab @ethzurich.bsky.social.
How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)
23.07.2025 11:40 —
👍 141
🔁 37
💬 5
📌 3
I'm thrilled to share that our story is now out in Science Advances! 🎉 We use quantitative imaging to map the mito central dogma, define translation hubs in the mitochondrial matrix, and show that they're replaced by Mitochondrial Stress Bodies (MSB) when mtRNA processing is perturbed 1/4
18.04.2025 22:56 —
👍 97
🔁 36
💬 8
📌 1
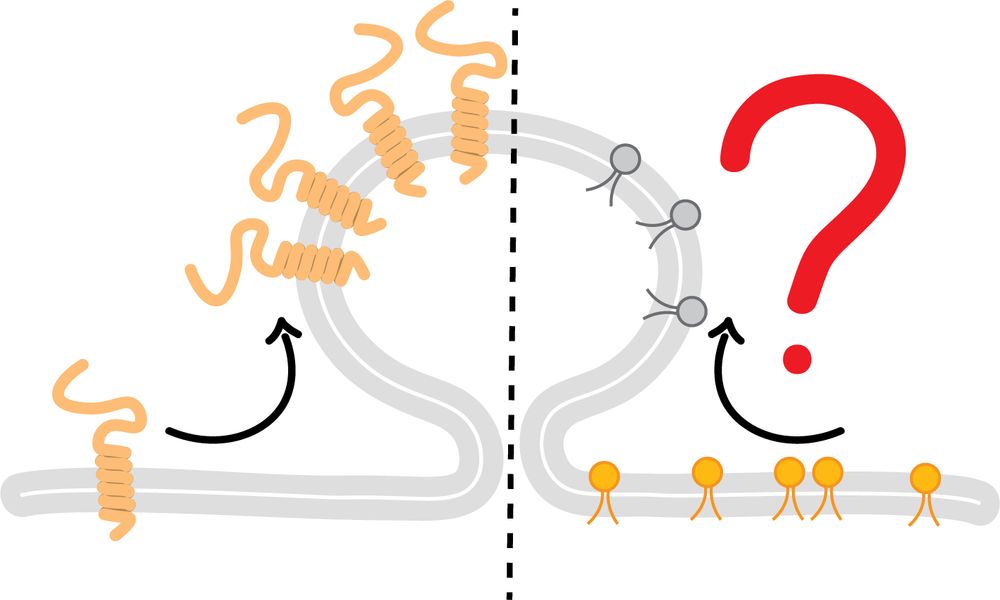
Are lipids actively sorted during clathrin mediated endocytosis like proteins? @mathilda95.bsky.social addresses this key open question together with our collaborators from the Honigmann and Modes labs using a new Lipid-STED workflow.
www.biorxiv.org/content/10.1...
10.03.2025 11:56 —
👍 114
🔁 38
💬 2
📌 9
Hey #chemsky and beyond, with all that is going on, I am curious what expensive, high margin molecules we could provide to the community for free. What things do you use all the time, cost way too much, and would make a difference? Alexa Fluor 488 NHS? Other dyes? Other molecule?
01.03.2025 13:57 —
👍 68
🔁 30
💬 7
📌 1

A proper ☃️ for a legit snow day in HOUSTON!!? Mother Nature, never let them know your next move!
21.01.2025 21:50 —
👍 2
🔁 0
💬 0
📌 0

Structured protein domains enter the spotlight: modulators of biomolecular condensate form and function
Biomolecular condensates are membraneless organelles that concentrate proteins and nucleic acids. One of the primary components of condensates is mult…
It's about time we have a conversation about the role of structured protein domains in form and function of bimolecular condensates. Proud of graduate student Nate Hess for taking the lead on synthesizing these ideas and identifying important gaps in the field!! www.sciencedirect.com/science/arti...
17.01.2025 22:05 —
👍 39
🔁 13
💬 3
📌 1
I am excited to report the preprint of my postdoc work in the @olzmannlab.bsky.social! We have discovered the first lipid droplet quality control pathway with broad implications for lipid physiology and diseases associated with LD accumulation and oxidative stress! #lipidtime
shorturl.at/2HqFd
08.01.2025 13:46 —
👍 64
🔁 15
💬 6
📌 0
Is chromatin ordered or disordered? It all depends on the linker DNA length.
Check our latest work with Mike Rosen and Sy Redding. We explore how changes in linker DNA length (as small as 1 bp) fine-tune chromatin structure, between order and disorder, and the properties of chromatin droplets
24.12.2024 09:48 —
👍 114
🔁 38
💬 6
📌 3

Gearing up for our lab’s holiday party! Grilling pizza, meats and veggies!!
19.12.2024 17:42 —
👍 3
🔁 0
💬 1
📌 0
Great thread from @levin-ferreyra.bsky.social about her latest work in the lab, out today in @emboreports.bsky.social ! 👇
13.12.2024 20:22 —
👍 6
🔁 1
💬 0
📌 0

Title and abstract of the memoriam in the Journal of Cell Biology in the link
In September the world lost one of the great biologists of our time: Joe Gall. He bridged the transition from the histology era of cell biology to our molecular present, contributing a remarkable set of insights in his 9 decades 1/n 🧪
rupress.org/jcb/article-...
16.11.2024 13:28 —
👍 220
🔁 76
💬 6
📌 3
Thank you!!
14.11.2024 01:44 —
👍 1
🔁 0
💬 0
📌 0
Great list!! I just migrated over. Could you please add me?
13.11.2024 15:37 —
👍 1
🔁 0
💬 1
📌 0
Lastly, I want to thank the editors and reviewers @jcellbiol.bsky.social for a constructive review that significantly improved the paper. They also provided an excellent example of peer review to the trainees in our lab, many of which have never published before this paper. Very grateful 🙏
13.11.2024 14:54 —
👍 4
🔁 1
💬 0
📌 0
13/Please check out their beautiful paper where they developed a complementary approach to evaluating interactions within condensates using FLIM fluorescence cross-correlation spectroscopy www.nature.com/articles/s41...
13.11.2024 14:54 —
👍 1
🔁 0
💬 1
📌 0
12/Eleonora Perego and Giuseppe Vicidomini (vicidominilab.github.io) gave excellent insight into what we were seeing, and provided us the opportunity to demonstrate our approaches translate to data captured from different FLIM platforms.
13.11.2024 14:54 —
👍 0
🔁 0
💬 1
📌 0
11/A huge bottleneck in our quest to understand our data and automate our pipelines came from an inability to convert raw TCSPC data from Leica LasX to G and S coordinates.. Our solution came from an open source GUI (FLUTE) from Chiara Stringari's Lab. pmc.ncbi.nlm.nih.gov/articles/PMC...
13.11.2024 14:54 —
👍 0
🔁 0
💬 1
📌 0
10/Josh Marcus spearheaded our Python scripts for a wavelet filter to resolve subtle fluorescence lifetime differences, and machine-learning segmentation of condensate lifetime clusters! Leyla Fahim wrote our automated FLIM-FRET scripts! github.com/LeeLabBCM
13.11.2024 14:54 —
👍 0
🔁 0
💬 1
📌 0










