🚨🚨 News from the @castello-lab.bsky.social and @shabazlab.bsky.social. Our work, led by Louisa Iselin, reveals pervasive changes in the RNA-bound proteome induced by interferon. #RNA, #immunity, #interferon, #host-virus www.biorxiv.org/content/10.1...
03.06.2025 22:01 — 👍 18 🔁 6 💬 1 📌 0
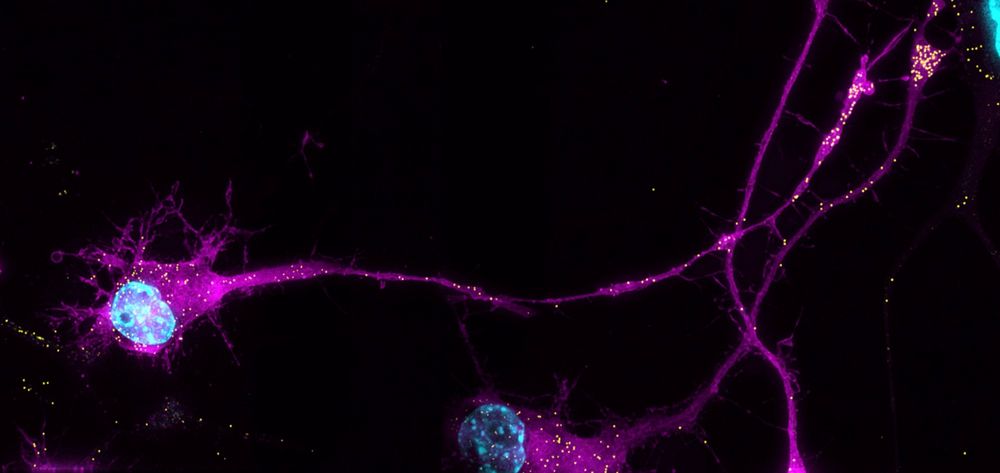
Excited to share our latest work! We asked if and how TDP-43 regulates RNA transport in neurons and if this is misregulated in ALS. We found that it works to keep RNAs *out* of neurites, and that loss of TDP-43 activity results in specific RNAs aberrantly accumulating there.
03.03.2025 15:55 — 👍 114 🔁 36 💬 6 📌 2
Thank you, Alfredo! 🥳
18.02.2025 11:39 — 👍 1 🔁 0 💬 0 📌 0

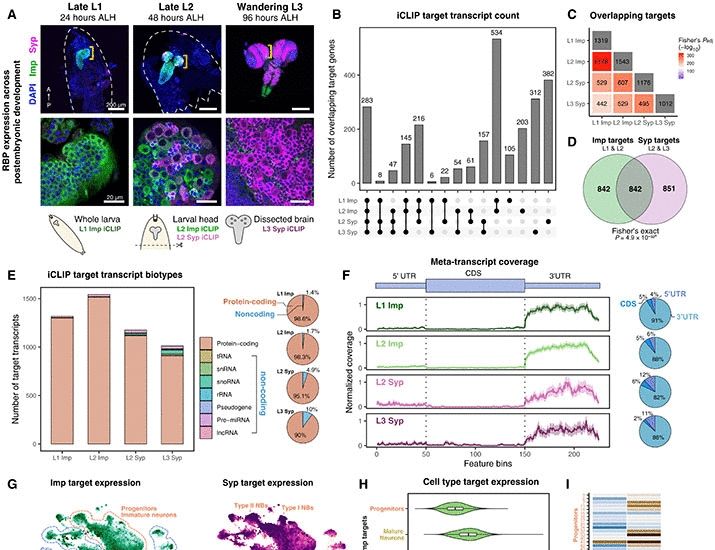
Imp/IGF2BP and Syp/SYNCRIP temporal RNA interactomes uncover combinatorial networks of regulators of Drosophila brain development
RNA binding protein combinations control neuronal identity through a network of temporal regulators.
Jeff Lee et al paper from my lab is out now
www.science.org/doi/10.1126/...
I am very grateful to Jeff, Nile's and Tamsin for all the hard work on this tour de force paper.
Jeff deserves more credit than me for masterminding it and the outstanding data analysis: 2 RBPs over 3 developmental times.
12.02.2025 23:18 — 👍 35 🔁 13 💬 1 📌 0

Following our recent publication, here’s a striking collage of developing fly brain! 🧠✨ Highlighting the dynamic expression of RNA-binding proteins Imp (green) and Syp (magenta) www.science.org/doi/10.1126/...
10.02.2025 17:42 — 👍 4 🔁 1 💬 0 📌 0
Grateful to Niles, Tamsin, @ilandavis.bsky.social, and our amazing collaborators for their support in this journey! 🙌
10.02.2025 17:31 — 👍 0 🔁 0 💬 0 📌 0

Imp/IGF2BP and Syp/SYNCRIP temporal RNA interactomes uncover combinatorial networks of regulators of Drosophila brain development
RNA binding protein combinations control neuronal identity through a network of temporal regulators.
🧠🔬How do RNA-binding proteins shape brain development? Our latest study reveals how Imp & Syp form combinatorial networks to regulate Drosophila neurogenesis. Excited to share this work in #ScienceAdvances!
📜 Read more: www.science.org/doi/10.1126/... #ilandavislab #RNA #neurogenesis
10.02.2025 17:30 — 👍 7 🔁 1 💬 1 📌 0
RNA biology of embryonic development and disease #heart #ribosome #translational_control #RBP #splicing @UK_Frankfurt Alum:@salkinstitute @UcSandiego
https://kurianlab.com/
RNAholic, Scientist, interested in architectural roles, packaging and binding codes
#RNA, #splicing, #condensates
Wellcome ECA Fellow in the Harding Lab at the #WCIP. Interested in nutrient regulation in Toxoplasma. She/Her. 🏳️🌈
Lab Account | PI: @bbrennan83 | 🔬 bunyavirus/vector interactions at @CVRinfo.bsky.social🏴🇬🇧🇱🇹🇹🇭🇪🇺🏳️⚧️🏳️🌈
Cell states & cell fates in zebrafish development
Lab Head since 2020
Imaging, Genetics, scRNAseq
Represents my personal opinions 🏳️🌈
Postdoc @mndoxford.bsky.social @ndcnoxford.bsky.social
working on protein biomarkers of ALS/MND and extracellular vesicles.
Evolutionary Virologist based at the University of Tokyo 🇯🇵
💻🦠🏳️🌈
World citizen, Time and Variation in Brain Development, football, cheese, wine, single malt!
Visualizing RNA regulation in living cells, one molecule at a time.
Human and viral RNAs.
Hubrecht Institute, Oncode Institute and dept. of Bionanoscience, TU Delft
Prof TT Uni Bonn & University Hospital Bonn | RNA & ribosome & biochemistry & innate immunology | Postdoc Barna Lab Stanford | PhD Stoecklin Lab DKFZ ZMBH Uni Heidelberg | triplet | https://www.leppeklab.org
RNA, Glycans, Space, Renewable Energy, and all technologies making lives more interesting.
PI at Boston Children’s Hospital SCP
Assistant Professor at Harvard SCRB
Researcher @UniofOxford in single molecule imaging of gene regulation at @RobKlose lab
I paint: https://aleksanderszczurek.wixsite.com/ats-art
Group leader at Max Perutz Labs interested in protein quality control
Head of Genome Biology Dept @EMBL,
Scientist, Principal investigator, Professor
Exploring genome regulation during development,
and everything to do with enhancers.
3D genome, chromatin topology,
cell_fate, embryonic Development,
Single Cell genomics
News, commentary and research coverage from the international monthly journal publishing the highest quality of work in all areas of neuroscience.
https://www.nature.com/neuro/
RNA biology, gene regulation, nervous system function and development
Just another LLM. Tweets do not necessarily reflect the views of people in my lab or even my own views last week. http://rajlab.seas.upenn.edu https://rajlaboratory.blogspot.com
🧬 Join our world-class courses, conferences, and workshops at the forefront of molecular life science and its applications. 👩🏻🔬 www.embl.org/events/ 🔬
RNA biologist at University of Colorado Anschutz Medical Campus http://www.taliaferrolab.com