The May Institute, if you don't know, combines the full workup of generating mass spec proteomics data all the way to doing the statistics.
This year has a few new modules, like FragPipe with @nesvilab.bsky.social and @fcyucn.bsky.social, so even if you've attended in the past, check it out!
20.01.2026 23:33 — 👍 8 🔁 3 💬 0 📌 0

A New Detailed Mass Offset Search in MSFragger for Improved Interpretation of Complex PTMs pubs.acs.org/doi/10....
---
#proteomics #prot-paper
14.01.2026 15:01 — 👍 11 🔁 3 💬 0 📌 0
So glad to see this online today! With FragPipe and the many other Koina APIs, we wanted to democratize deep learning, especially for those without access to expensive GPUs. Major kudos to my co first author Ludwig for setting up the server. I encourage ML developers to put their models on Koina
12.11.2025 04:37 — 👍 10 🔁 1 💬 0 📌 0
This project started 5 years ago. It led us to add isotope-labeling support to #FragPipe/#IonQuant. Since then, the tools have grown so much and are now widely used in #Chemoproteomics.
Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social
30.10.2025 14:15 — 👍 15 🔁 5 💬 0 📌 0

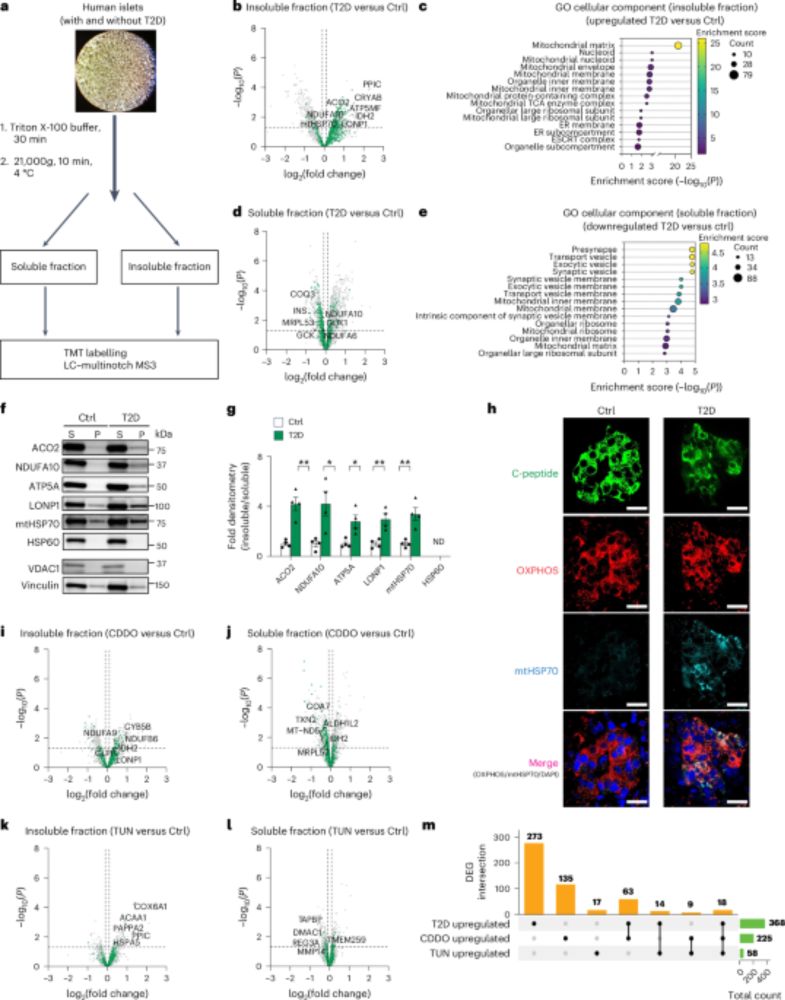
This work led by Elena Levi-D'Ancona, a recent PhD graduate in our lab, was our first cover and only possible due to our amazing team and outstanding collaborators, including @nesvilab.bsky.social and Orian Shirihai, and funding from NIDDK, Breakthrough T1D, and the VA! Thank you! 🤗🙌 2/fin
08.10.2025 20:28 — 👍 3 🔁 1 💬 1 📌 0
Start Page: /home/software/Skyline/events/2025 Webinars/Webinar 26
Proteomics Webinar: DIA with FragPipe, DIA-NN, and Skyline
Presenters: Eduard Sabidó and Brendan MacLean
When: Tuesday, September 16, 8am (Pacific Time)
Register Now ... skyline.ms/project/home...
#massspec #proteomics
15.09.2025 08:26 — 👍 33 🔁 16 💬 0 📌 0
exact quote is "this site is being blocked due to DOJ Bulk Sensitive Data Regulations as the site is hosted in a Geo-Fenced country". I think replication of the data would be the only option for us in the foreseeable future.
10.09.2025 18:01 — 👍 2 🔁 0 💬 2 📌 0
The University of Michigan now blocks the iProX database, which is a part of the PRIDE consortium of mass spec data repositories. All requests to unblock were denied. Any other US universities in a similar situation? There is a lot of valuable MS proteomics data there no longer accessible to us.
10.09.2025 17:44 — 👍 9 🔁 3 💬 1 📌 0


It was a great pleasure to teach #FragPipe at the Biological Proteomics for Beginners workshop at #UCSD, sponsored by Thermo Fisher Scientific. We had a fantastic group of grad students, postdocs, and professors. Yes, I even got to teach UCSD professors how to analyze bottom-up proteomics data 😁
10.09.2025 14:52 — 👍 18 🔁 1 💬 1 📌 0
Dan Polasky is indeed a perfect teammate, and not only in our lab but also apparently as a … player in Kubb. I also want to use this opportunity to publicly congratulate Dan for being promoted to Research Assistant Professor starting September 1st!
28.08.2025 21:11 — 👍 11 🔁 0 💬 0 📌 0

PSManalyst: A Dashboard for Visual Quality Control of FragPipe Results
FragPipe is recognized as one of the fastest computational platforms in proteomics, making it a practical solution for the rapid quality control of high-throughput sample analyses. Starting with version 23.0, FragPipe introduced the “Generate Summary Report” feature, offering .pdf reports with essential quality control metrics to address the challenge of intuitively assessing large-scale proteomics data. While traditional spreadsheet formats (e.g., tsv files) are accessible, the complexity of the data often limits user-friendly interpretation. To further enhance accessibility, PSManalyst, a Shiny-based R application, was developed to process FragPipe output files (psm.tsv, protein.tsv, and combined_protein.tsv) and provide interactive, code-free data visualization. Users can filter peptide-spectrum matches (PSMs) by quality scores, visualize protease cleavage fingerprints as heatmaps and SeqLogos, and access a range of quality control metrics and representations such as peptide length distributions, ion densities, mass errors, and wordclouds for overrepresented peptides. The tool facilitates seamless switching between PSM and protein data visualization, offering insights into protein abundance discrepancies, samplewise similarity metrics, protein coverage, and contaminants evaluation. PSManalyst leverages several R libraries (lsa, vegan, ggfortify, ggseqlogo, wordcloud2, tidyverse, ggpointdensity, and plotly) and runs on Windows, MacOS, and Linux, requiring only a local R setup and an IDE. The app is available at (https://github.com/41ison/PSManalyst.
It's now properly published. If you want to easily check important characteristics of your data before diving into complicated statistics, check out PSManalyst.
PSManalyst: A Dashboard for Visual Quality Control of FragPipe Results | Journal of Proteome Research pubs.acs.org/doi/10.1021/...
15.08.2025 19:45 — 👍 24 🔁 7 💬 2 📌 0

Profiling the proteome-wide selectivity of diverse electrophiles
Targeted covalent inhibitors are powerful entities in drug discovery, but their application has so far mainly been limited to addressing cysteine residues. The development of cysteine-directed covalen...
Interested in the proteome-wide selectivity of diverse electrophiles?
The proteomics data for our study on this headed by @pzanon.bsky.social are now public on @pride-ebi.bsky.social: www.ebi.ac.uk/pride/archiv...
Full story: chemrxiv.org/engage/chemr...
#ChemBio #ChemSky #ChemicalProteomics
12.08.2025 09:50 — 👍 21 🔁 8 💬 0 📌 0
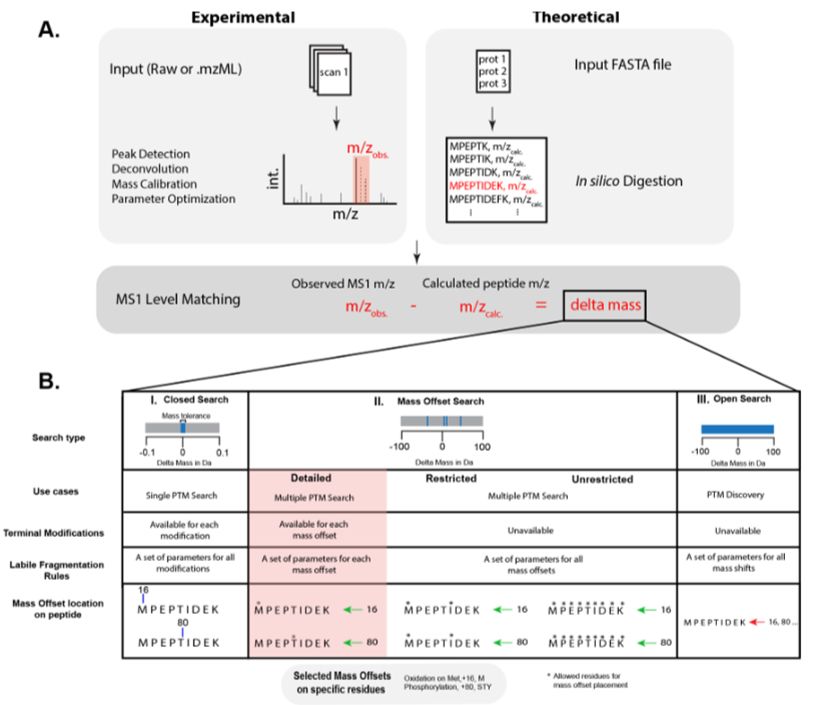
Conventional proteomics searches struggle with many modifications and fully open searches may be difficult to interpret. We introduce a "detailed" mass offset search in #MSFragger boosting interpretability and localization especially in complex cases (e.g. FPOP data): www.biorxiv.org/content/10.1...
01.08.2025 21:33 — 👍 34 🔁 6 💬 0 📌 0
Have you compared FragPipe on i9 vs comparably priced AMD threadripper? I put a request to our IT to get me one but they do not support core imaging of AMD machines. I am still recommending a good Xeon or i9 to folks who want a standard desktop to run FragPipe on a < $5k machine but want to test AMD
24.07.2025 19:09 — 👍 1 🔁 0 💬 1 📌 0
Don't tell PD, but I have quietly switched all of my analyses to Fragpipe. What an extremely powerful software. I'm often amazed at the sheer quantity of information I can get using Fragpipe. Thanks to everyone who recommended it.
11.07.2025 20:49 — 👍 12 🔁 2 💬 2 📌 0
I miss Twitter a bit. With its outrageous commercials and all those gorgeously looking “Lucy564” people who seemed to wanted to follow me in droves. Ok, not really. I do occasionally check Bluesky but I only see posts from a few (mostly same) people. Lack of critical mass?
14.06.2025 17:03 — 👍 5 🔁 0 💬 2 📌 0

Wow, a record breaking number of the Nesvizhskii lab members attending #ASMS2025! 9 posters, 3 evening workshops, and one Bioinformatics Hub on #FragPipe. Plus multiple collaborative posters with other groups. See you in Baltimore! PS. Below is our recent group photo, including all those attending
30.05.2025 19:45 — 👍 35 🔁 8 💬 0 📌 1
The power of #MSFragger open search! “we applied the mass-tolerant search engine MSfragger and found that phosphorylation as well as ubiquitination were well preserved after XDNAX. To our great interest, we found an additional modification of 321 Da occurring only in the irradiated SILAC channel”
23.05.2025 12:41 — 👍 14 🔁 2 💬 0 📌 0

Targeted proteomics: Advanced tools for biomedical research
Targeted proteomics technologies, and specially data-independent acquisition techniques, have revolutionized the landscape of proteomic research in the last decade offering researchers unprecedented …
Don't miss out! Applications still open for EMBO Practical Course on Targeted proteomics: Advanced tools for biomedical research in Barcelona, Spain, 5 – 10 October 2025
Abstract submission & registration deadline: 15 May
meetings.embo.org/event/25-tar...
#EMBOtargetedProteomics #EMBOevents 🧪
05.05.2025 09:53 — 👍 6 🔁 3 💬 0 📌 2
If there any #Sciex decision makers here on Bluesky - I urge you to reconsider. Skyline/Proteowizard support is not only important for your customers using these tools, but it also benefits other bioinformatics efforts that depend on these tools.
15.05.2025 01:24 — 👍 25 🔁 9 💬 0 📌 1
The tools remain free to academics under academic license. There has not been a discussion up to this point about changing that. Thanks to all our users, academic (now many thousands) and commercial, for their support!
09.05.2025 11:49 — 👍 12 🔁 0 💬 0 📌 0
Thank you so much for inviting us!
09.05.2025 01:30 — 👍 1 🔁 0 💬 0 📌 0
No, you still need to get a pre-release version from us. Email us please
09.05.2025 01:20 — 👍 0 🔁 0 💬 0 📌 0
Staff Scientist in Mass spectrometry platform @fmp-berlin.de. Interested in cross-linking mass spectrometry and interactomics.
Assistant Professor of Medicine
MSc, PhD, FAHA
https://cics.bwh.harvard.edu
Mass spectrometry
Proteomics
Opinions are my own, they should be yours too.
Translational Neuroproteomics Lab | Understanding mechanisms of protein (dis)aggregation in neurodegenerative diseases | Views = own
Proteomics lab in Sheffield (UK) studying protein interactions and PTMs in health and disease with a particular interest in neuroproteomics.
Mass Spectrometrist, mother, cat servant, dog helper, and sometimes other things
Postdoc in Küster lab at Technical University of Munich (TUM), Germany. Protein-nucleic acid interactions & photo-Xlinking proteomics | RNA | mass spectrometry | RNA-interacting proteomes | DNA-interacting proteomes | nucleotide-crosslinked peptides
Mass spectrometry for quantitative proteomics. The Olsen group at the Center for Protein Research, University of Copenhagen.
Professor, Founder and Director of the Centre for Targeted Protein Degradation at the University of Dundee. We develop molecules that induce protein interactions and degradation
Senior Scientist at Arena Bioworks in Chemical Proteomics (Cambridge, MA). Formally of The Francis Crick Institute.
We productized proteomics services and tranformed the field. Here to discuss all things proteomics and occasionally stir the pot :)
At MSAID, I am very lucky to work with incredibly intelligent people at the interface between mass spectrometry-based proteomics and artificial intelligence.
Science, glycobiology, mass spectrometry, coffee. Assoc. professor @unimelb. Former ARC future fellow. he/his
Data Science Coordinator @SMOC (Swiss Multiomics Center (smoc.ethz.ch))
We are a chemical biology lab at the Nuffield Department of Medicine at the University of Oxford.
Talus Bio is a venture-backed early-stage biotechnology startup company working on drug development for gene regulators.
Chemical biologist, basketball aficianado, proud homebody. Personal account.