Applications are open for the @crg_eu PhD Programme! 20 fully funded positions — including one in our group through the Evolutionary Medical Genomics ITN.
Join us to develop deep generative models of cross-species data to tackle open questions in disease genetics.
www.crg.eu/en/content/t...
23.10.2025 11:00 —
👍 11
🔁 9
💬 0
📌 3
We also made some improvements with genomic language model, Evo 2, but in this case the interpretation was less clear. See the preprint for more details. Code for using LFB will made available shortly. 10/10
26.05.2025 17:30 —
👍 2
🔁 0
💬 0
📌 0
This provides evidence that better fitness estimation can be achieved at negligible computational cost by bridging the gap between likelihood and fitness at inference time. 9/n
26.05.2025 17:30 —
👍 2
🔁 0
💬 1
📌 0
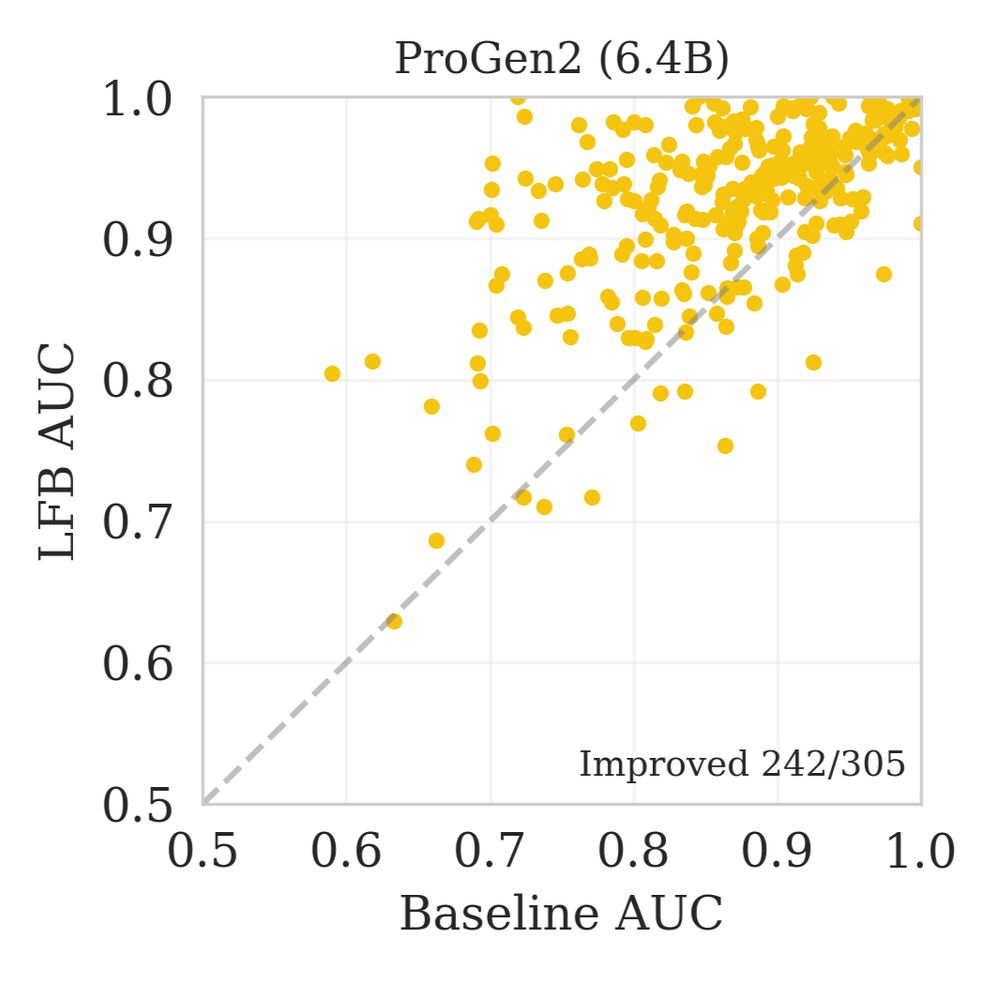
We show a scatterplot of ROC-AUCs for each gene, calculated separating benign and pathogenic labelled variants with either usual or LFB fitness estimation
This trend held across DMS assay types and mutational depth, and also on prediction of clinical variants. 8/n
26.05.2025 17:30 —
👍 1
🔁 0
💬 1
📌 0
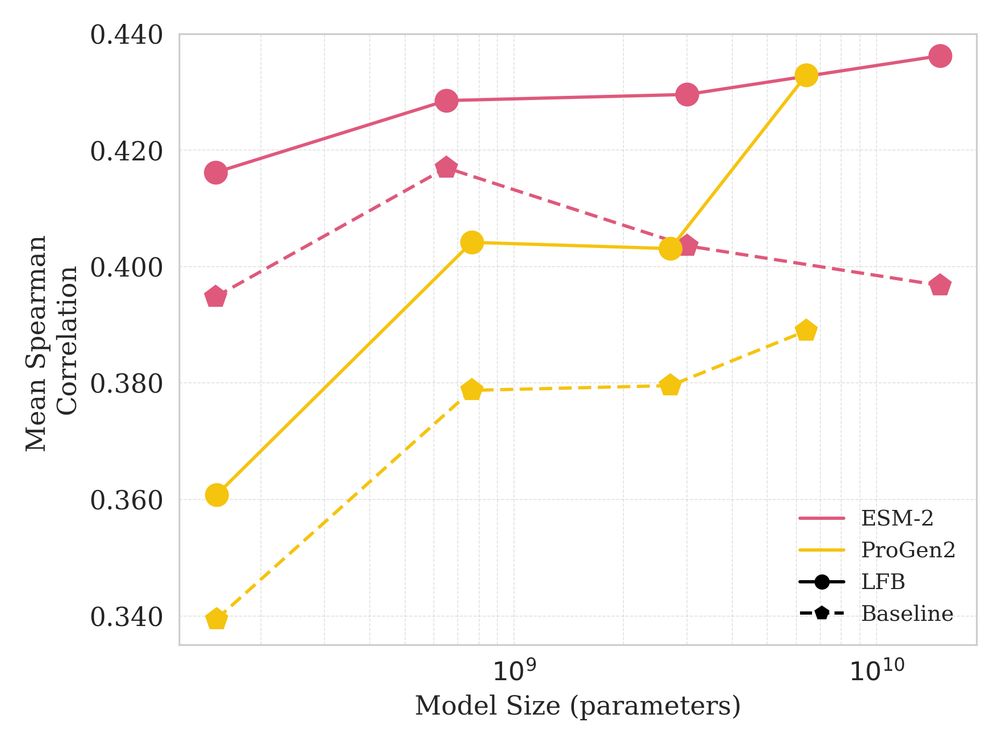
We show a plot of Model Size vs Mean Spearman Correlation across the DMS datasets from ProteinGym for ESM-2 and ProGen2 model families both with and without the LFB estimation.
On ProteinGym, LFB provided significant improvements across model classes and sizes and we saw that larger better fit models provided better predictions in general.
proteingym.org 7/n
26.05.2025 17:30 —
👍 2
🔁 0
💬 1
📌 1
We found under an Ornstein–Uhlenbeck model of evolution that our LFB should be lower variance than the standard estimate by marginalising the effect of drift. 6/n
26.05.2025 17:30 —
👍 2
🔁 0
💬 1
📌 0
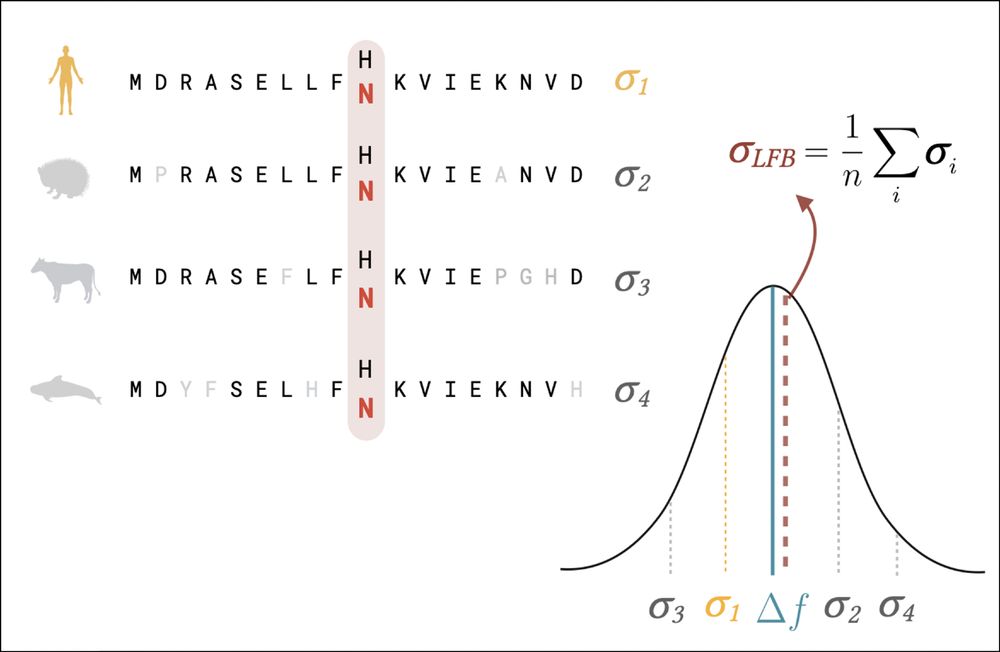
We show a schematic of the LFB estimate where by averaging over predictions for a variant applied to other related sequences, we produce an score which should be closer to the true change in fitness.
We tried a simple strategy — averaging predictions over sequences under similar selective pressures to effectively reduce the impact of unwanted non-fitness related correlations — likelihood fitness bridging (LFB). 5/n
26.05.2025 17:30 —
👍 1
🔁 0
💬 1
📌 0
We wondered whether we might be able to improve predictions from existing models without any further training. 4/n
26.05.2025 17:30 —
👍 1
🔁 0
💬 1
📌 0
Non-identifiability and the Blessings of Misspecification in Models...
Misspecification is a blessing, not a curse, when estimating protein fitness from evolutionary sequence data using generative models.
Weinstein et al show that better fit sequence models can perform worse at fitness estimation due to phylogenetic structure:
openreview.net/forum?id=CwG...
And in practice we are seeing that pLMs don’t improve with lower perplexities:
openreview.net/forum?id=UvP... www.biorxiv.org/content/10.1... 3/n
26.05.2025 17:30 —
👍 1
🔁 0
💬 1
📌 0
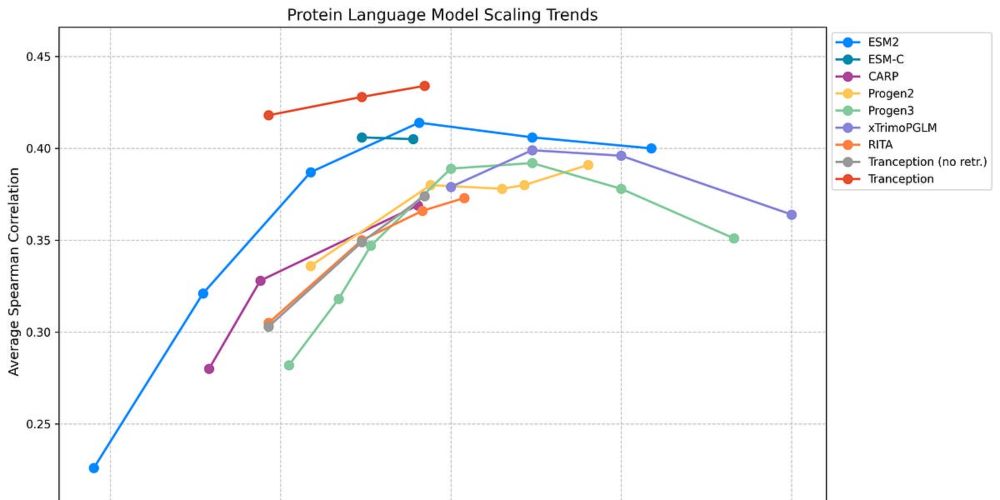
Have We Hit the Scaling Wall for Protein Language Models?
Beyond Scaling: What Truly Works in Protein Fitness Prediction
Protein language models are showing promise in variant effect prediction - but there’s emerging evidence likelihood based zero shot fitness estimation is breaking down. See this excellent summary from @pascalnotin.bsky.social: pascalnotin.substack.com/p/have-we-hi... 2/n
26.05.2025 17:30 —
👍 5
🔁 0
💬 1
📌 0
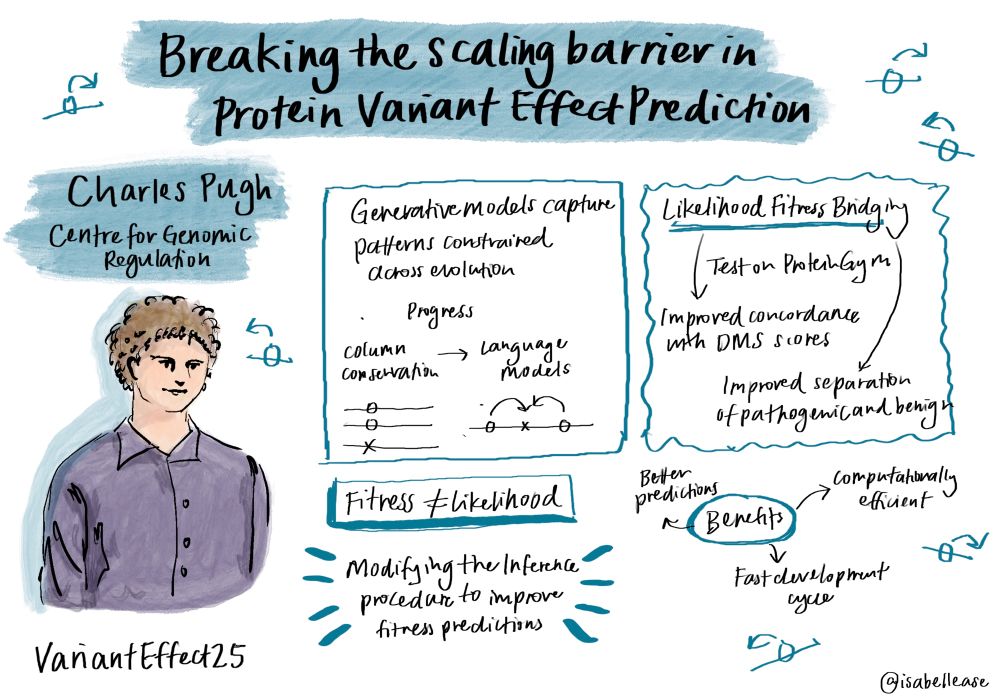
@cwjpugh.bsky.social at #VariantEffect25
22.05.2025 10:29 —
👍 19
🔁 8
💬 0
📌 0
Three BioML starter packs now!
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
03.12.2024 03:27 —
👍 149
🔁 58
💬 16
📌 6

Thanks Charlie for opening the PhD Symposium! Many thanks to everyone involved in its organisation. #CRGPhDSymp2024
28.11.2024 09:10 —
👍 7
🔁 4
💬 0
📌 0






