Super excited to get this out. This collab started a few years ago and is the first paper from it. Here, with experimental and computational approaches we:
1. establish that cell villages can be just as accurate (one might argue more accurate!) than arrayed-based designs
bsky.app/profile/bior...
29.09.2025 16:47 — 👍 40 🔁 21 💬 2 📌 0
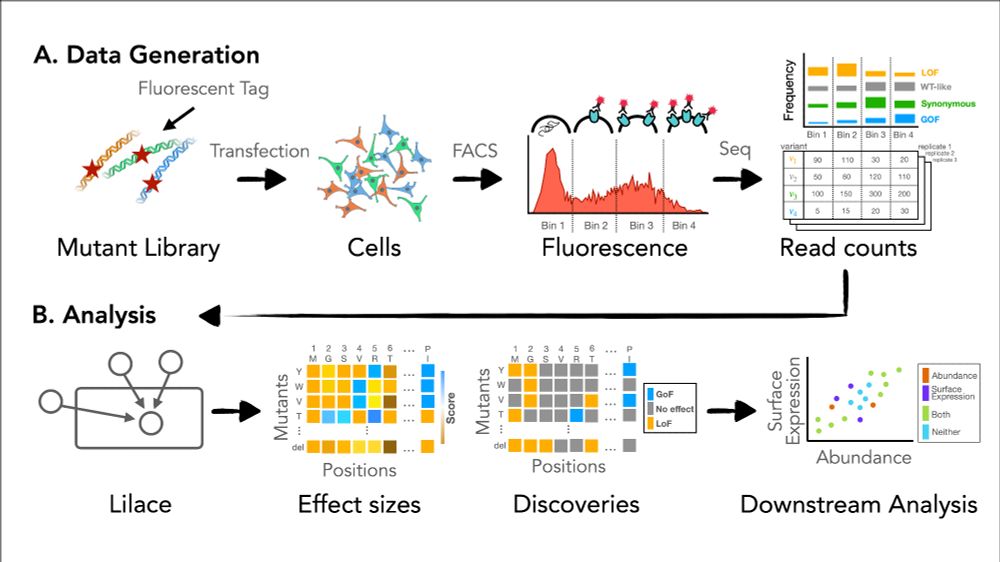
figure 1 of Lilace paper detailing the data collection and analysis process to indicate where the Lilace model fits in
Statistical assessment of score uncertainty is also becoming especially critical as DMS experiments scale to more conditions, phenotypes, and proteins. We hope Lilace will help address these challenges and enable more reliable functional interpretation of FACS-based DMS data! (3/3)
30.06.2025 16:03 — 👍 3 🔁 0 💬 0 📌 0
Coupling DMS experiments with FACS enables simultaneous measurement of a quantitative phenotype (e.g. protein abundance) across thousands of mutations in a protein. However, modeling FACS as a phenotype can be challenging due to its unique multidimensional nature and experimental noise. (2/3)
30.06.2025 16:03 — 👍 4 🔁 0 💬 1 📌 0

Accurate variant effect estimation in FACS-based deep mutational scanning data with Lilace
Deep mutational scanning (DMS) experiments interrogate the effect of genetic variants on protein function, often using fluorescence-activated cell sorting (FACS) to quantitatively measure molecular ph...
Check out our new preprint on Lilace, a statistical tool for scoring FACS-based deep mutational scanning experiments! Lilace directly models the shift between variant fluorescence distributions and provides score uncertainty estimates to better assess reliability and reproducibility. (1/3)
30.06.2025 16:03 — 👍 18 🔁 5 💬 1 📌 3
NYT bestselling author of EMPIRE OF AI: empireofai.com. ai reporter. national magazine award & american humanist media award winner. words in The Atlantic. formerly WSJ, MIT Tech Review, KSJ@MIT. email: http://karendhao.com/contact.
avid grocery shopper | clarion 2025 | vp 2024 | codex | words in clarkesworld + khoreo
repped by @allegramartsch.bsky.social
clairejiawen.com
2nd year computational neuro PhD student at Boston University (advisors Cynthia Bradham & Gabe Ocker)
Mathematically modeling embryonic neurodevelopment
I write trippy scifi & mathfiction.
Ignyte Award Finalist 2025
6 stories in Clarkesworld
speculative fiction author still excited by having her own wikipedia page. won some things. lost some things. wrote that hole story.
debut novel SUBLIMATION from Tor 6/2/26.
http://isabel.kim < website
Wow if True < listen wherever pods are cast
Climate scientist; ocean carbon cycle and climate solutions. Professor, University of Hawaiʻi at Mānoa; Visiting Faculty, Arizona State University. https://linktr.ee/david_ho
i write at @theverge.com. Portland, OR.
Publishing reviews and commentaries across the fields of genetics and genomics. Part of Springer Nature and Nature Portfolio.
https://www.nature.com/nrg/
Official account of the RNA Society | We're a non-profit international scientific society with more than 1800 members dedicated to fostering research and education in the field of RNA science.
https://www.rnasociety.org
Nuclear events
Narratives in chromatin
By haiku, of course
EPIGENETIC HULK SMASH PUNY GENOME. MAKE GENOME GO. LOCATION: NOT CENTROMERE, THAT FOR SURE
The real jbouie. Columnist for the New York Times Opinion section. Co-host of the Unclear and Present Danger podcast. b-boy-bouiebaisse on TikTok. jbouienyt on Twitch. National program director of the CHUM Group.
Send me your mutual aid requests.
We’re an independent, nonprofit organization that works side by side with consumers to create a fair and just marketplace.
staff writer @theatlantic.com and senior fellow @snfagora.bsky.social. author of GULAG, IRON CURTAIN, RED FAMINE, TWILIGHT OF DEMOCRACY and AUTOCRACY INC
https://linktr.ee/anneapplebaum
Study genetic conflicts professionally. Try to avoid conflicts in personal life (with mixed results).
Fred Hutch Basic Sciences,
UW Genome Sciences,
HHMI.
Posting in a personal capacity. My posts don’t reflect my employers’ opinions.
Professor, UW Biology / Santa Fe Institute
I study how information flows in biology, science, and society.
Book: *Calling Bullshit*, http://tinyurl.com/fdcuvd7b
LLM course: https://thebullshitmachines.com
Corvids: https://tinyurl.com/mr2n5ymk
he/him
xRNA therapeutics | Head of Biofoundry & RNA by Design Leader @ Centillion | Views my own | #RNAsky #RNAtx #RNArocks
Scientific Project Manager in the NINDS Office of Research Quality
Loves high-quality research, particularly in neuroscience and RNA biology
Working to change the culture of science
Views are my own
"RNA is the love language of the genome."
Science person, RNA fan, lab account over at areslab.bsky.social
Interested in RNAs that must adopt two or more distinct structures to function. Also interested in mechanisms of intron gain during genome evolution.
UC San Diego Biochemistry studying RNA localization and mitochondria
🥼 structural biology & RNA biochemistry
👩🏻🔬 postdoc @ MSKCC
🎓 Emory University '21
🏫 University of Richmond '16


