Very exited to be awarded my first SNSF Project Funding as the project PI. Huge thanks to @snsf.ch and the very supportive comments from the reviewers! The grant will support my project to decode human brain development and disease with AI-driven regulatory network modeling
08.10.2025 14:36 — 👍 4 🔁 0 💬 0 📌 0
Excited to launch our AlphaGenome API goo.gle/3ZPUeFX along with the preprint goo.gle/45AkUyc describing and evaluating our latest DNA sequence model powering the API. Looking forward to seeing how scientists use it! @googledeepmind
25.06.2025 14:29 — 👍 219 🔁 82 💬 5 📌 10
Modular integration of epithelial, microbial, and immune components to study human intestinal biology. Amazing team project with Rubén López-Sandoval, Marius F. Harter, Qianhui Yu, Matthias Lütolf, Mikhail Nikolaev, and Nikolche Gjorevski! biorxiv.org/content/10.1...
09.05.2025 12:54 — 👍 17 🔁 6 💬 1 📌 0
Great to be part of the team! And this would not be the end of our efforts on exploring and characterizing the organoid systems
03.05.2025 06:14 — 👍 4 🔁 0 💬 0 📌 0

CellFlow enables generative single-cell phenotype modeling with flow matching
High-content phenotypic screens provide a powerful strategy for studying biological systems, but the scale of possible perturbations and cell states makes exhaustive experiments unfeasible. Computatio...
Yay, we built a thing! With @dominik1klein.bsky.social Daniil, Aviv Regev, Barbara Treutlein @graycamplab.bsky.social @fabiantheis.bsky.social we use flow matching to enable generalised sc phenotype modeling. From cytokine screens to fate programming and organoid engineering tinyurl.com/3xhju7db
18.04.2025 10:19 — 👍 35 🔁 9 💬 1 📌 0

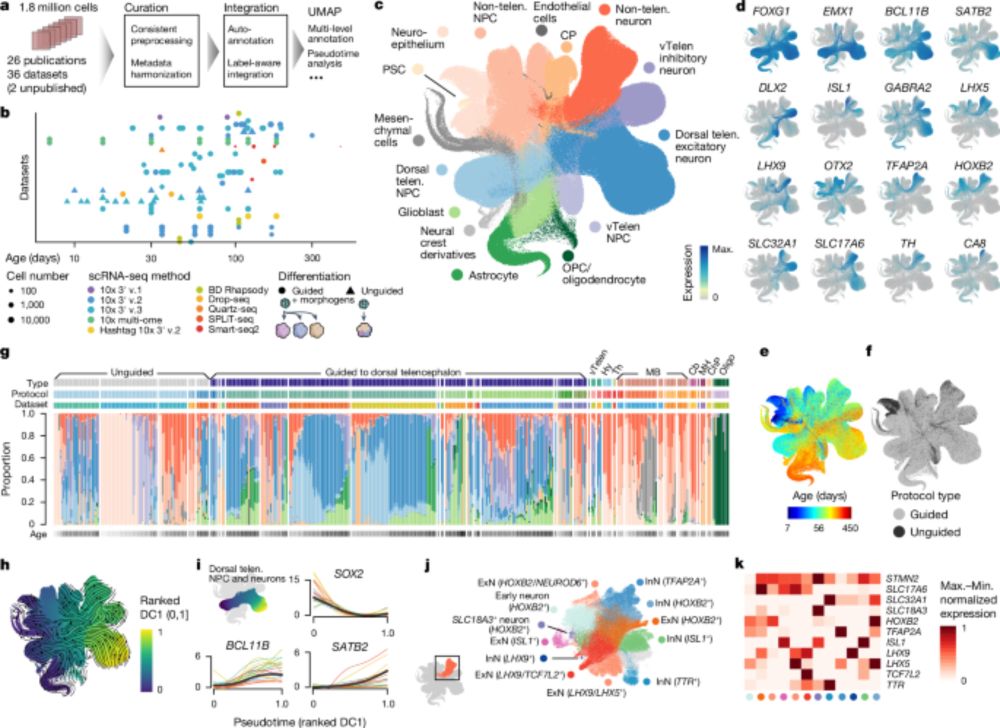
Multi-omic human neural organoid cell atlas of the posterior brain
Patterning of the neural tube establishes midbrain and hindbrain structures that coordinate motor movement, process sensory input, and integrate cognitive functions. Cellular impairment within these s...
New preprint from the @graycamplab.bsky.social & Treutlein labs, offering a single-cell multi-omic atlas and morphogen screen of human neural organoid models of the midbrain and hindbrain (#cerebellum) 🧪🧫🧬🧠
Project led by
@nazbukina.bsky.social @zhisonghe.bsky.social @hsiuchuanlin.bsky.social
21.03.2025 08:38 — 👍 12 🔁 2 💬 0 📌 0
It was really great to work with @nazbukina.bsky.social @hsiuchuanlin.bsky.social and all people involved in this project! It is cool to see how we can learn about posterior brain development from organoids and how we can extend the organoid protocols from existing ones!
24.03.2025 14:08 — 👍 6 🔁 0 💬 0 📌 0
Looking forward to all the exciting projects and studies from your lab in the coming years!
23.11.2024 17:37 — 👍 1 🔁 0 💬 0 📌 0

Excited to announce that I will start my group at @crg.eu in Barcelona next April! We will decode and engineer cell fates using stem cell models, perturbation screening and single-cell multiomics. Looking forward to the new chapter ahead!
23.11.2024 16:41 — 👍 59 🔁 11 💬 6 📌 2

Glad that our neural organoid atlas is out @Nature! Many interesting analyses for in vitro-in vivo comparisons beyond neural organoid setting - here eg fig 4e, where we visualize on primary reference which brain regions are not yet covered by organoids. Exciting for exp design!
22.11.2024 17:25 — 👍 6 🔁 2 💬 0 📌 0
Hi thanks for making the pack! Can you add me to the pack too please?
21.11.2024 08:12 — 👍 1 🔁 0 💬 1 📌 0
Thanks for making the pack, can you add me to the pack too?
21.11.2024 07:47 — 👍 2 🔁 0 💬 1 📌 0
PhD Student and Animal Fact Enthusiast in the Domcke Lab, DMLS UZH. She/her.
Where the future begins. 🚀🔬 One of the world’s leading universities for technology & natural sciences. Posts in both English and German.
www.ethz.ch
Postdoc @ethz.ch Barbara Treutlein Lab | PhD @Lund university Malin Parmar Lab | Human brain development, single cell sequencing, cell replacement therapy 🧠🧫🧬
Assistant Professor at at Stanford Biology & Stem Cell Institute. Studying human brain development through genomics and proteomics. Website: www.liwanglab.org
The German Stem Cell Network (GSCN) aims at creating synergies between all areas of basic and applied stem cell research and to provide an interface between science, education, politics and society as a whole. More information here: www.gscn.org
'Cells & Development' journal (formerly 'Mechanisms of Development'). Official journal of the International Society of Developmental Biology isdb.bsky.social
A scientific journal publishing cutting-edge methods, tools, analyses, resources, reviews, news and commentary, supporting life sciences research. Posts by the editors.
Chief Editor of Nature Biomedical Engineering. Ph.D. Previously at Nature Methods. @rita_strack on the place formerly known as twitter.
ISDB is a non-profit scientific association that promotes the study of developmental biology
PhD Candidate @CRG.eu with @larsplus.bsky.social | Single Cell Tech Dev & Functional Genomics
Reprogramming cells for fun and to create embryo models in the tropical Greater Bay Area
https://www.jose-silva-lab.com/ 🇵🇹🇨🇳🇬🇧
Guangzhou/Cantāo 🌴☀️, China中国
Postdoc @bhadurilab.bsky.social
Human brain development + single-cell omics
Our mission: To create comprehensive reference maps of all human cells—the fundamental units of life—as a basis for both understanding human health and diagnosing, monitoring, and treating disease. https://www.humancellatlas.org
Stem cell and developmental biologist. Modeling embryo development. Assistant Professor @Westlake University.
https://en.westlake.edu.cn/faculty/lizhong-liu.html
A preprint highlights service run by the biological community and supported by The Company of Biologists (@biologists.bsky.social).
Research, news, and commentary from Nature, the international science journal. For daily science news, get Nature Briefing: https://go.nature.com/get-Nature-Briefing
ELLIS PhD student @HelmholtzMunich, Student Researcher @Apple. Interested in ML, Single-Cell Genomics, and People.