Happy to share the 2nd part of my PhD @natsmb.nature.com!
Using CRISPR screens, we uncover the TF-RE wiring underlying Xist regulation. Since the preprint, we've added a titration of Xist, showing that XCI is directly dependent on Xist levels.
Check the Bluetorial by @eddaschulz.bsky.social 👇
06.10.2025 17:04 — 👍 18 🔁 3 💬 1 📌 0


@jengreitz.bsky.social l & my lab want to co-hire a computational biologist/biostatistician with project management expertise to help map the regulatory code of the human genome and discover genetic mechanisms of disease.
Details below
careersearch.stanford.edu/jobs/computa...
Plz RT
19.08.2025 00:29 — 👍 55 🔁 60 💬 1 📌 1
This is not a HiC map! Ever wondered if multiple enhancers get activated simultaneously? We measured chromatin accessibility on thousands of molecules by nanopore to create genome-wide co-accessibility maps. Proud of @mathias-boulanger.bsky.social @kasitc.bsky.social Biology in the thread👇
18.08.2025 13:00 — 👍 111 🔁 34 💬 10 📌 0

CRISPR screen decodes SWI/SNF chromatin remodeling complex assembly www.nature.com/articles/s4...
31.05.2025 14:15 — 👍 6 🔁 2 💬 0 📌 0
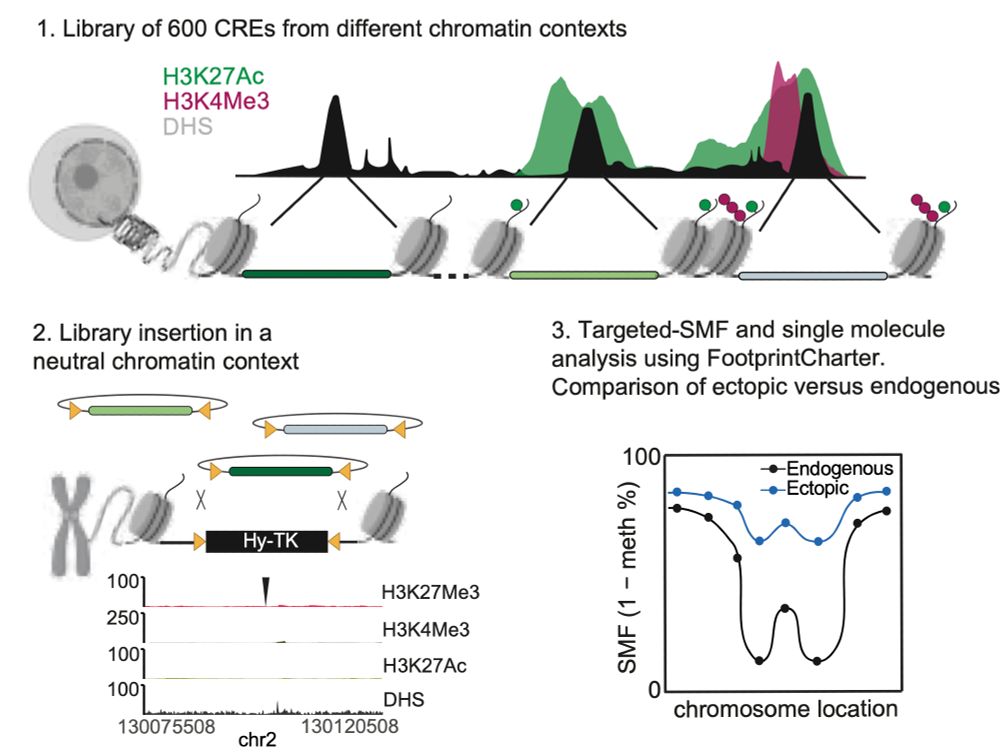
Happy to share the latest story from @arnaudkr.bsky.social's lab @embl.org! With @guidobarzaghi.bsky.social, we used Single Molecule Footprinting to quantify how often chromatin is accessible at enhancers after TF and chromatin environment changes! Check our preprint bit.ly/3XQMFxN + thread ⬇️ 1/11
08.04.2025 13:51 — 👍 77 🔁 33 💬 4 📌 2
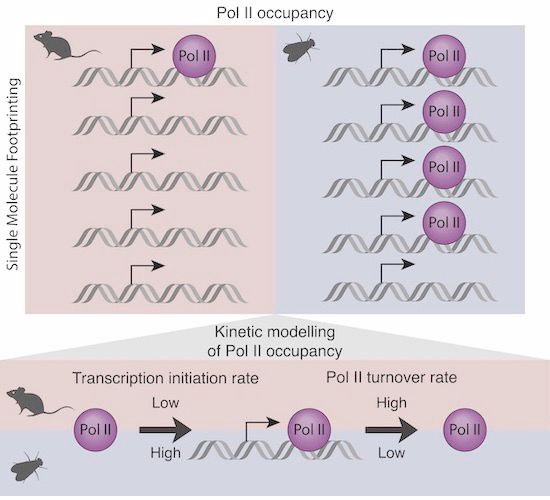
The importance of moving away from bulk! Occupancy of Pol II at promoters is dramatically different between fly and mouse cells! When looking single molecule! Proud of the team! @kasitc.bsky.social @molinalab.bsky.social doi.org/10.1038/s443...
02.04.2025 08:47 — 👍 115 🔁 37 💬 2 📌 1
It’s official 🎉 I am excited to share that I will start my lab this spring at the Max Planck Institute for Immunobiology and Epigenetics @mpi-ie.bsky.social in beautiful Freiburg 🤩🗻☀️ I am looking forward to new collaborations and working with the fantastic community in Freiburg & its surroundings!
16.01.2025 18:38 — 👍 182 🔁 31 💬 28 📌 5

A throwback to last month's 'Quantitative biology to molecular mechanisms' conference – time to introduce the poster prize winners! #EMBLOmics
A round of applause for:
🏅 Max Trauernicht
🏅 @ingridpelaez.bsky.social
🏅 Honorine Destain
🏅 Óscar García Blay
Read on 👉🏻 s.embl.org/omx24-01-blog
@embl.org
19.12.2024 14:22 — 👍 9 🔁 4 💬 0 📌 0


I attended the ‘Quantitative Biology to Molecular Mechanisms’ conference at EMBL Heidelberg last week and had the incredible honor of winning one of the Poster Prizes! 🏆
A big thanks to the organizers and all the amazing speakers who shared their inspiring research with us. ✨
#EMBLOmics
27.11.2024 09:26 — 👍 18 🔁 5 💬 0 📌 1
PhD Student in Luca Giorgetti's Lab (@lucagiorgetti.bsky.social) - Genome architecture - Microscopy - Image analysis - Biophysics
PhD Student with @marcbuhler.bsky.social at @fmiscience.bsky.social
Master's in Heidelberg, thesis with @karsten-rippe.bsky.social
Editorial assistant at the @nightsciencepod.bsky.social
gene regulation, chromatin, nuclear organisation, synthetic biology
Incoming Group Leader @molgen | Postdoc in the Wysocka Lab at @ Stanford. @ EMBO, @ CancerResearch, and @ Leading Edge fellow.
Gene regulation | Transposons | Human embryo development
Scientist & serial-punner I Chromatin dynamics & gene regulation in early development I MSc. @reNEW | Ph.D. @embl | 🇩🇰 🇮🇹 |
Scientist at the German Cancer Reseach Center in Heidelberg. We develop genome engineering tools for functional genomics. #CRISPR #Drosophila #openscience
ML for regulatory genomics. PhD student @ Gagneurlab
johahi.github.io
Learning from algorithms that learn from data about gene regulation.
Scientist/PI Hubrecht Institute. Exploring gene regulation in early development and cancer. Single-cell spatial genome organization & epigenomics
Exploring the interplay between gene regulation and tissue-tissue interactions in stem cell-based embryo models.
Cancer systems biologist. I'm poking at cancer cells to model how they work in details. #perturbation #model #highthroughout #targetedtherapy
ORCID: 0000-0002-1663-2075
Genetics, bioinformatics, comp bio, statistics, data science, open source, open science!
Postdoctoral researcher at @anshulkundaje.bsky.social Machine learning #ML, gene regulatory networks #GRN, single cell and spatial #omics.
Previously at @saezlab.bsky.social
Developer of https://decoupler.readthedocs.io/
Incoming group leader @mpi-mg, Berlin
Chromatin tracing, epigenetics, development. Equity and advocacy in academia.
Senior Bioinformatician / data analyst at @EMBL GeneCore - Ph.D in health biology and genomics | Former postdoc at @EMBL Krebs lab - PhD student at @igmm
Our laboratory seeks to understand how chromosome structure relates to genome functions
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
Assistant Professor, UBC school of Biomedical Engineering. Trying to enable personalized medicine by solving gene regulatory code.
News from the Gagneurlab@TUM -- To understand the genetic basis of gene regulation and its implication in diseases.
https://www.cs.cit.tum.de/cmm
VP, Genentech
Associate Professor at the Allen School of Computer Science and Engineering, University of Washington (on leave)








