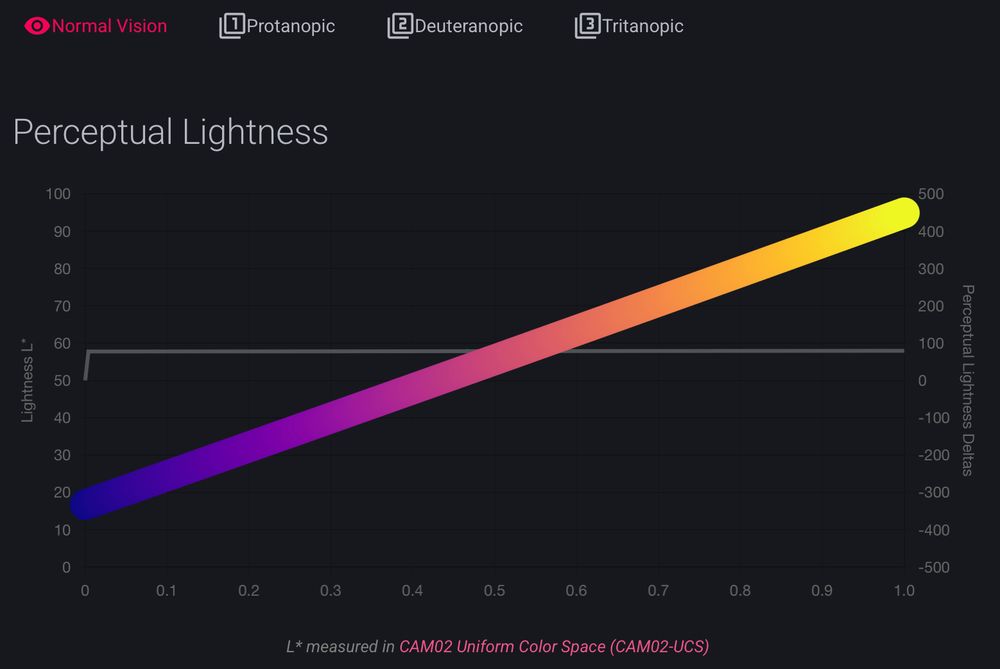
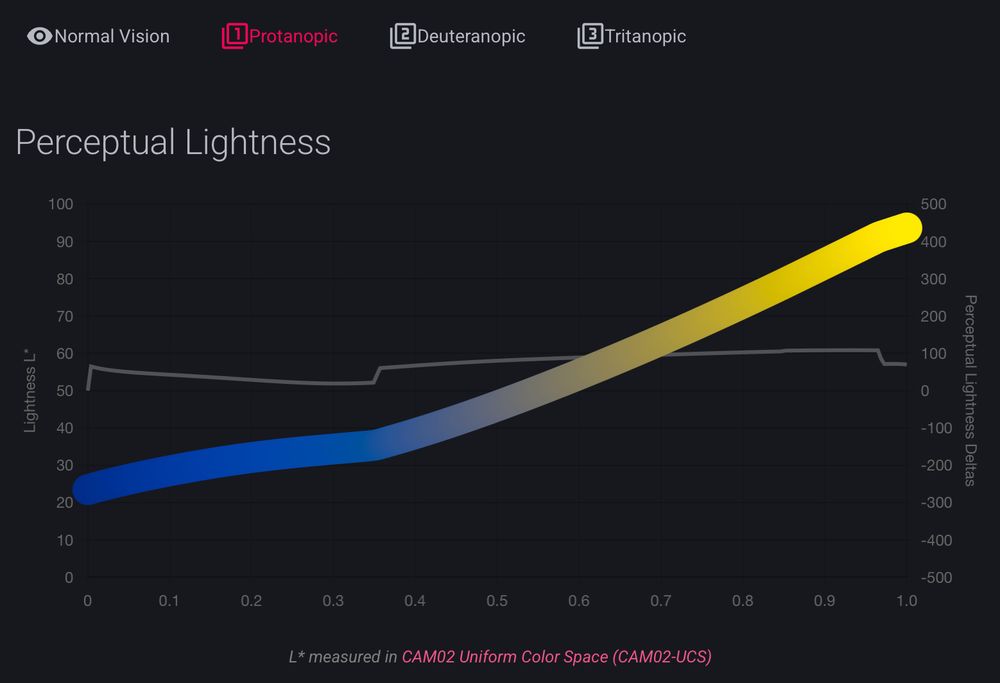
it was already fun to look around the documentation for cmap (cmap-docs.readthedocs.io)
but @grosoane.bsky.social just added a very cool feature to toggle between color vision deficiencies. Watch your perceptually uniform color maps gain some kinks 😂
as a mild deutan myself, I appreciate it! 🙏
05.07.2025 20:28 — 👍 19 🔁 6 💬 0 📌 0
Happy to officially introduce FilaBuster - a strategy for rapid, light-mediated intermediate filament disassembly. Compatible with multiple IF types, modular in design, and precise enough to induce localized filament disassembly in live cells.
www.biorxiv.org/content/10.1...
22.04.2025 01:02 — 👍 122 🔁 36 💬 2 📌 6
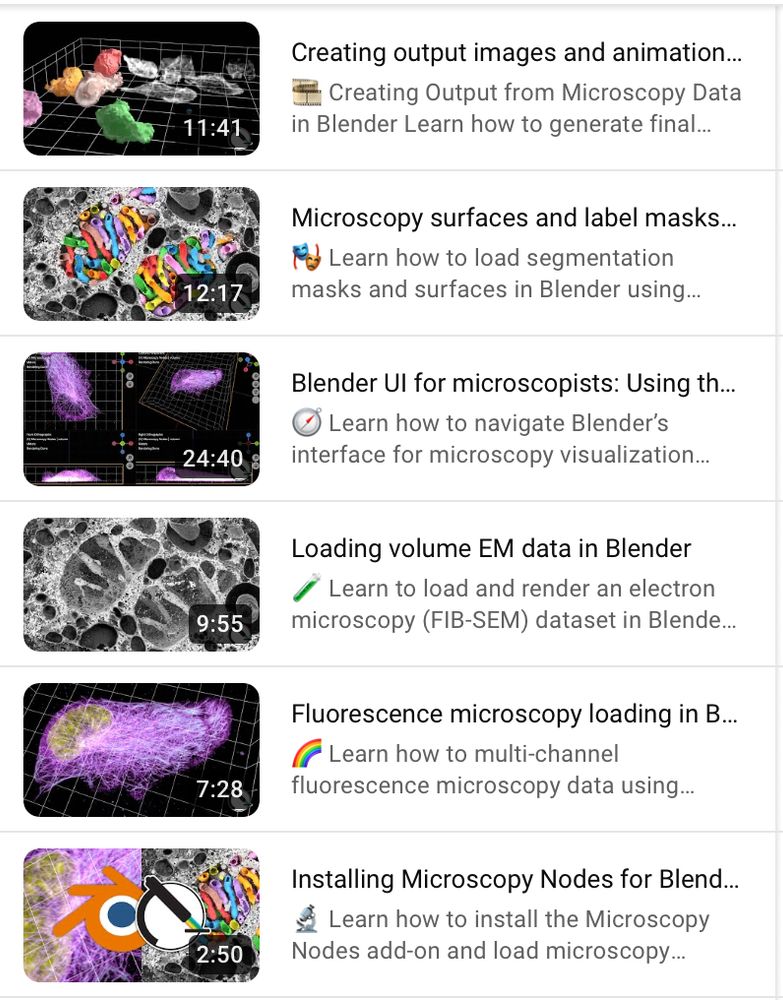
This is accompanied by a new set of tutorials showing exactly how you can start making beautiful microscopy visualizations in @blender.org!
Following along is also easy with the example OME-Zarr @openmicroscopy.org datasets 😄
Find the tutorials here: www.youtube.com/playlist?lis...
23.05.2025 13:13 — 👍 28 🔁 7 💬 0 📌 1
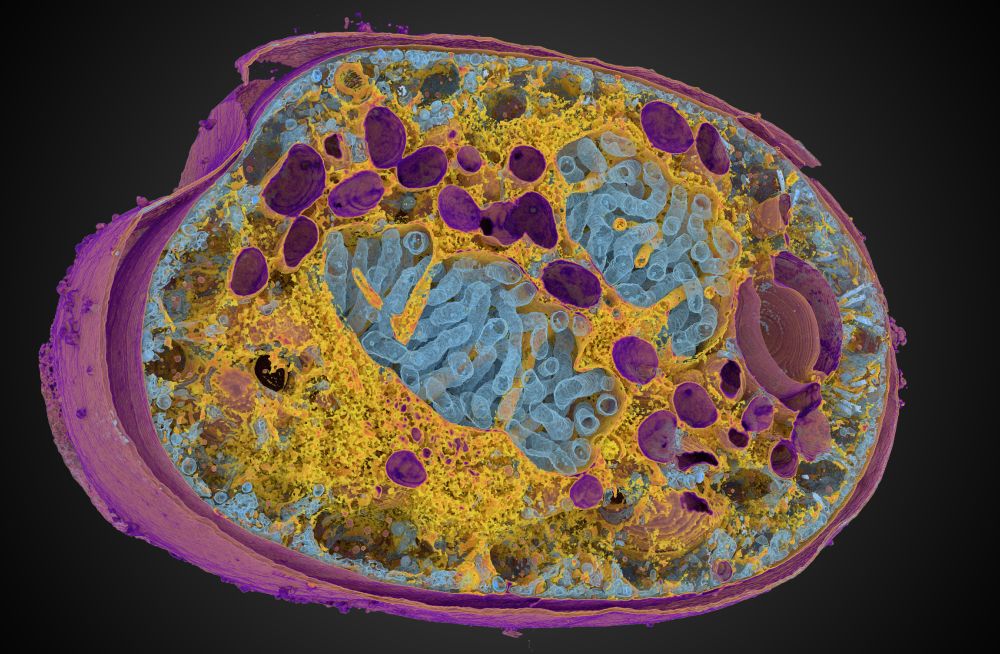
In the context of our @reviewcommons.org revision process, I'm happy to announce Microscopy Nodes v2.2.0!
This packs lots of new fun features, including new color management 🌈, clearer transparency handling 🫥, custom default settings 🔧 and more!
Preprint at doi.org/10.1101/2025...
23.05.2025 13:13 — 👍 87 🔁 31 💬 2 📌 2
ilastik - Download
New stable ilastik version 1.4.1 at www.ilastik.org/download!
Release Highlights:
- Support for OME-zarr (v0.4)
- New workflow - Trainable Domain Adaptation
- New object features based on Spherical Harmonics Feature, contributed by @grosoane.bsky.social.
- Native support for Apple Silicon
16.05.2025 10:45 — 👍 11 🔁 3 💬 1 📌 0
So excited to be a collaborator on this! We used @grosoane.bsky.social #microscopynodes to import tomo data to Blender before passing to Sensu for the beautiful animation.
03.04.2025 16:50 — 👍 7 🔁 1 💬 0 📌 0
YouTube video by Nemo Andrea
Procedural Microtubule | Blender 3.1 Geometry Nodes
It would only be appropriate to post about the #geometrynodes #microtubule here too. For a full introduction, I have prepared the world's least professional youtube video.
www.youtube.com/watch?v=AYUy...
07.03.2025 22:04 — 👍 19 🔁 3 💬 0 📌 1
Together with Christian Tischer, 3D rendering of multi-res microscopy data was added to MoBIE FIJI plugin using #BigVolumeViewer.
Check any remote datasets (like @dudinlab.bsky.social maps of protists using ExM) in the highest quality.
For full details read forum.image.sc/t/mobie-plus...
06.02.2025 20:55 — 👍 65 🔁 23 💬 0 📌 2
10 - Featured extension: Mastodon-Blender
Offers a bridge from Mastodon to Blender to create high quality 3D rendering of lineages and cell tracks over time:
(Tracking data of phallusia mammillata embryogenesis by Guignard et a. (2020). doi.org/10.1126/scie...)
30.01.2025 16:44 — 👍 17 🔁 3 💬 1 📌 0
And it also works in 2D 😁 Here we extract a circular projection instead of a spherical, and apply a 1D Fourier transform.
Here we can see how the channel distributions all align in the projection after we optically activate Rac1 in one part of the cell. 💡
With @jpassmore.bsky.social , Lukas Kapitein
30.01.2025 10:11 — 👍 2 🔁 0 💬 0 📌 0
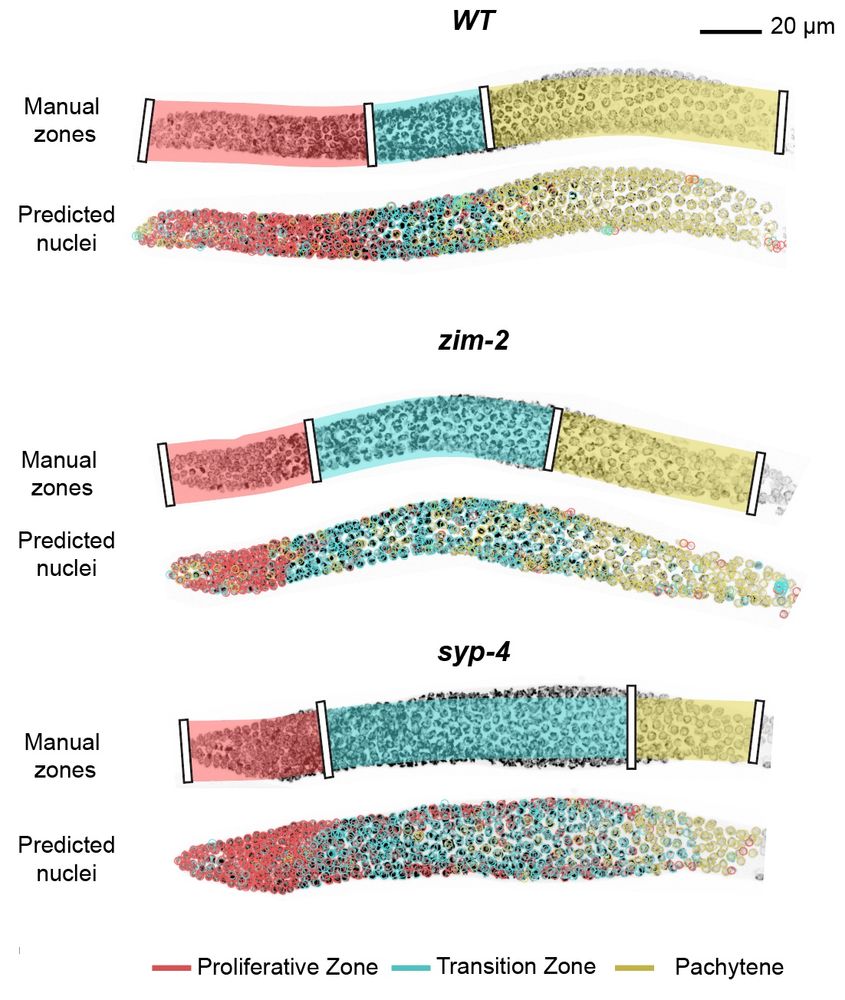
With the integration into the accessible toolkit ilastik (@ilastik-team.bsky.social) you can very easily include this in a quickly and interactively retrainable classification algorithm.
We show this for classification of meiotic zones in C. elegans :) 🪱
For the nerds there's also a Python API 😉
30.01.2025 10:11 — 👍 3 🔁 0 💬 1 📌 1
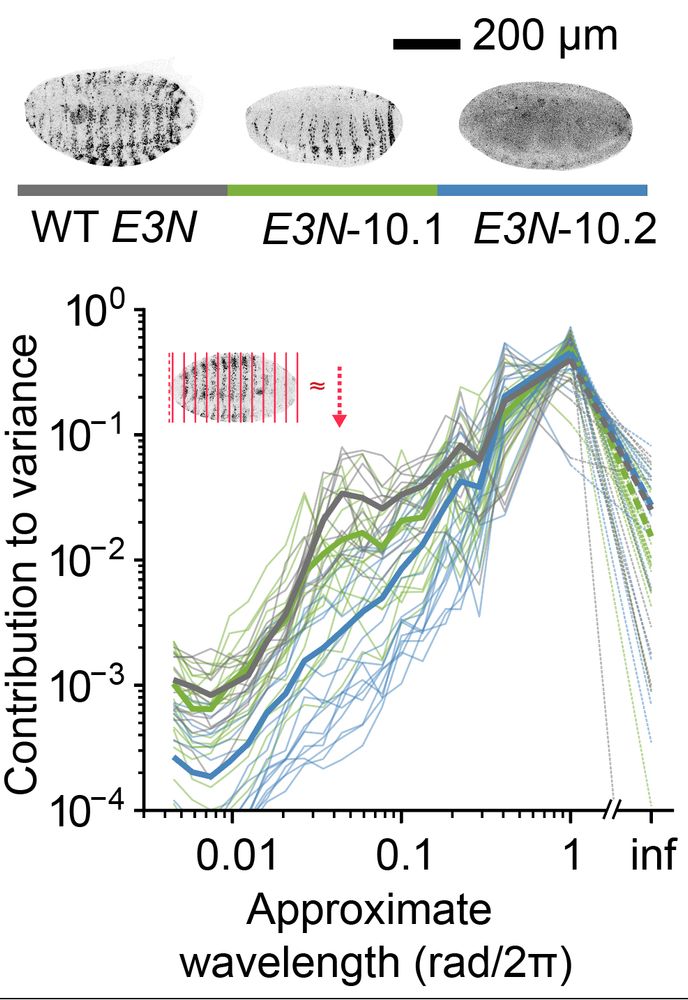
Quantifying distribution of intensity can be very helpful in many biological systems. For example, in D. melanogaster data, we extract a peak for the level of embryonic patterning in development. 🪰
In collaboration with
@ottilie.bsky.social, @timothyfuqua.bsky.social, @justinmcrocker.bsky.social
30.01.2025 10:11 — 👍 6 🔁 2 💬 1 📌 0
We then use spherical harmonics decomposition to quantify the variance in the spherical signal per angular wavelength.
This gives us a metric that shows how detailed the features in the spherical map are, giving a rotation-invariant quantification of distribution.
30.01.2025 10:11 — 👍 2 🔁 0 💬 1 📌 0
To quantify a segmented microscopy object, we project this to a spherical mean map.
This video is made with @blender3d.bsky.social geometry nodes and Microscopy Nodes, allowing the microscopy and method illustration in the same video :D
30.01.2025 10:11 — 👍 4 🔁 0 💬 1 📌 0
Now out in PLOS CB: Spherical Texture extraction! doi.org/10.1371/jour...
This method quantifies the intensity distribution in microscopy objects, and is implemented parameter-free in @ilastik-team.bsky.social object classification!
We show applications in cells, C. elegans and Drosophila! 😁
30.01.2025 10:11 — 👍 39 🔁 9 💬 1 📌 2
The API access is on the docket to improve! I was now mostly focused on the GUI, leaving the API only for changing Zarr data.
I was thinking to add utility functions that wrap the internals of Microscopy Nodes, but I'd love to discuss what's necessary! Can you open an issue on the GitHub page? 😁
15.01.2025 14:25 — 👍 2 🔁 0 💬 1 📌 0
To learn more, please check out my youtube tutorials at www.youtube.com/@oanegros which shows, among others, how to make this video!
The data here is a dinoflagellate from Mocaer et al. 2023, hosted publicly at EMPIAR-11399 (also shown in the first post here).
15.01.2025 13:39 — 👍 28 🔁 6 💬 1 📌 0
Microscopy Nodes loads data from a @zarr.dev bucket, making it easy to use open data archives that host Zarr data!
This is a mitotic cell from Walther et al. 2018, that's hosted publicly on the Image Data Resource 😱
15.01.2025 13:39 — 👍 10 🔁 2 💬 1 📌 0
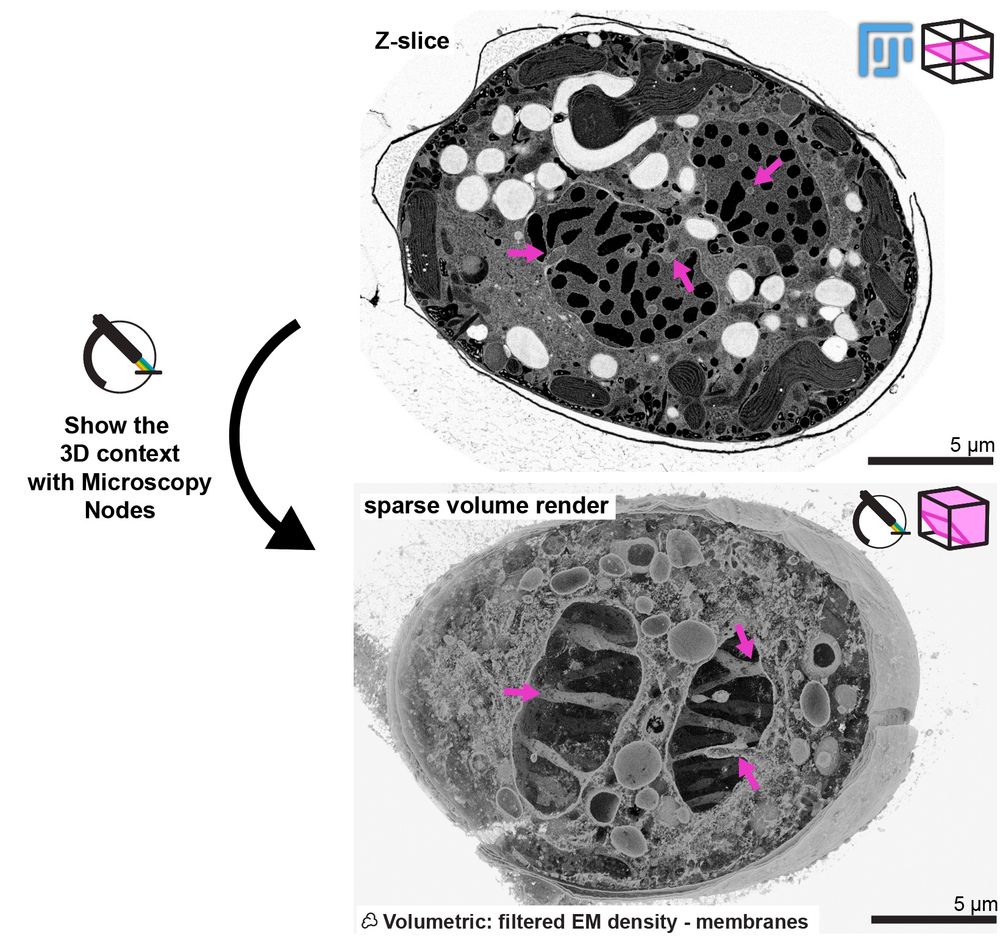
Here you can see how Microscopy Nodes can show the really cool phenotype of this mitotic dinoflagellate taken by @chandnibhickta.bsky.social!
With the sparse volume render in Blender you can see the network of tunnels through the nucleus that organize the spindle in its closed mitosis! 🤩
15.01.2025 13:39 — 👍 7 🔁 2 💬 1 📌 0
With Microscopy Nodes, you can play with large volumes (this ExM dataset from Granita Lokaj is 49 GB total) and integrate this with all the cool tools in Blender, such as this #geonodes model of the centriole, made by @banterlegroup.bsky.social !
15.01.2025 13:39 — 👍 32 🔁 6 💬 1 📌 1
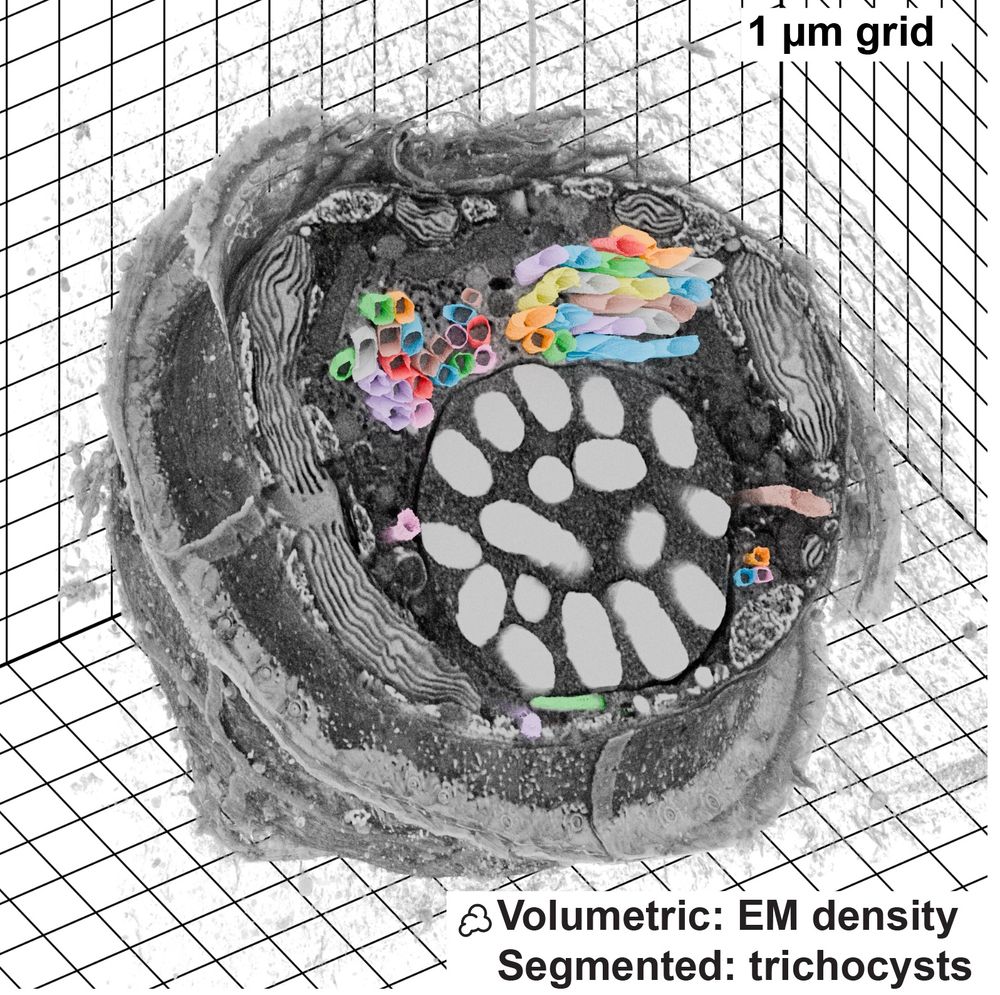
FIB-SEM dataset visualized with Microscopy Nodes, data from Mocaer et al 2023
Microscopy Nodes is now up on bioRxiv! 🚀
This is a Blender extension that seamlessly integrates and visualizes 3D microscopy data (TIF & @zarr.dev).
High-quality volume rendering for anyone, in both EM and fluorescence, regardless of computational expertise! 🔬
www.biorxiv.org/content/10.1...
15.01.2025 13:39 — 👍 334 🔁 124 💬 9 📌 12
That would be amazing! 😋
11.12.2024 07:57 — 👍 0 🔁 0 💬 0 📌 0

In honor of the first ever #MicroscopyNodes stickers arriving, I'm assembling my purse of open source bio(image) projects 😋. Which ones am I missing?😸
10.12.2024 09:23 — 👍 10 🔁 0 💬 3 📌 0
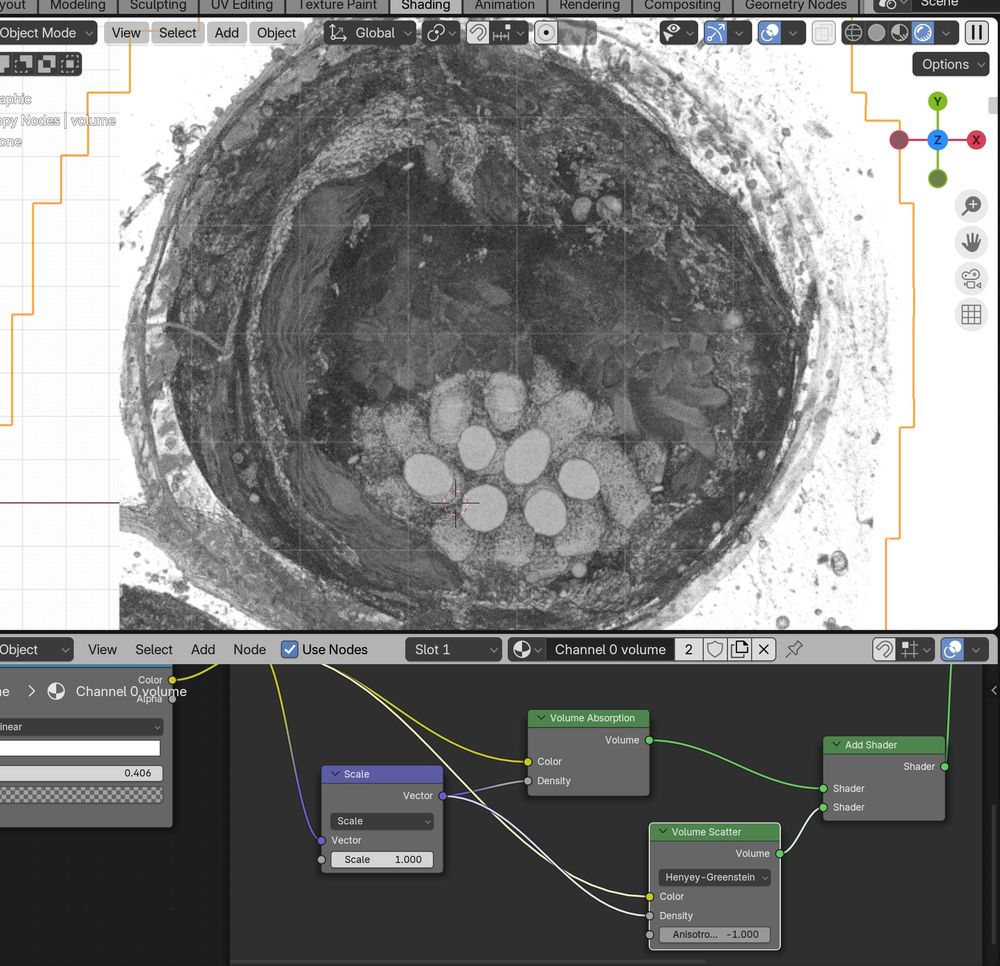
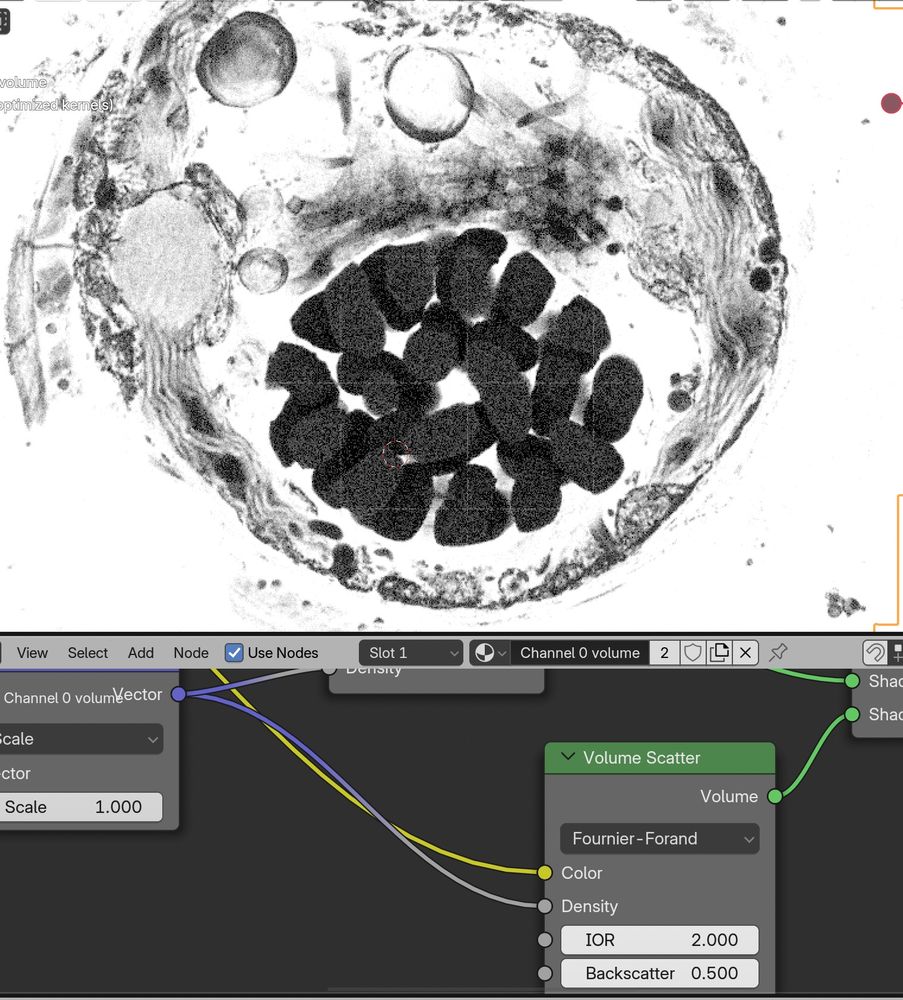
Messing around with the new #Blender 4.3 volume scatter modes with #microscopynodes, while at the #OME #Zarr hackathon! 😁🤩
Great info and work on the upcoming features of Zarr here :D Transforms, collections, and more! 🙏
20.11.2024 10:33 — 👍 28 🔁 7 💬 0 📌 0
Wow! Amazing video! Happy to see that Microscopy Nodes is useful to you 😋
I love that your render shows all the versatility of rendering microscopy data in Blender, that you don't often have access to in microscopy-specific implementations 😁🙏
18.11.2024 15:53 — 👍 11 🔁 0 💬 0 📌 0
And with OME-Zarr we can easily load data from public databases on the internet, such as the Bioimage Archive or IDR. Here is a beautiful mitosis dataset from Walther et al. (2018), hosted at uk1s3.embassy.ebi.ac.uk/idr/zarr/v0....
21.10.2024 08:30 — 👍 2 🔁 0 💬 0 📌 0

And with OME-Zarr loading, due to the pyramidal format, you can design your video at small-scale and only replace to full-scale at the final render!
(data from Granita Lokaj, @ s3.embl.de/microscopyno...)
21.10.2024 08:29 — 👍 17 🔁 3 💬 1 📌 0
With the new channel interface, you can define per channel features, such as whether to load as volume/isosurface/labelmask + channel names and more!
21.10.2024 08:29 — 👍 1 🔁 0 💬 1 📌 0
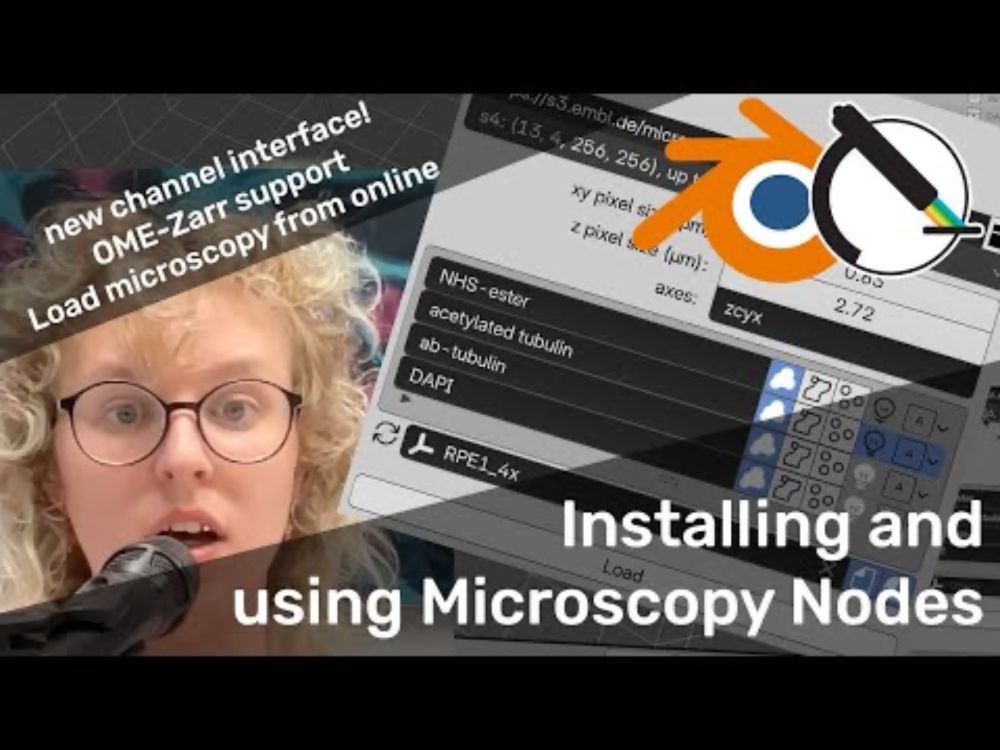
YouTube video by Oane Gros
Installing and using Microscopy Nodes in Blender
Microscopy Nodes 2.0 is out! This includes a new channel interface and OME-Zarr support! 🔬🎉
www.youtube.com/watch?v=ybQc...
21.10.2024 08:28 — 👍 21 🔁 5 💬 1 📌 1
Molecular animator, cell biologist, biochemist, science communicator. Head of the Animation Lab and Director of the Genetic Science Learning Center at the University of Utah, and writer of the RCSB PDB Molecule of the Month
Still seeking M. Ulder for joint author paper.
Microscopes ✅ cats ✅ combining the two ❌
Lab --> https://www.kcl.ac.uk/research/culley-group
Professor for Bioinformatics and Computational Oncology
University of Duisburg-Essen
Author of Snakemake, founder of Bioconda
Computational cryo-EM/ET 👨💻
Based in San Francisco 🌁
Love open source, mostly 🐍
I like cryoET, fluorescences microscopy and cell biology. Studying NPCs in the Beck Lab at MPIBP Frankfurt. MBL Physiology’23 alumn. (She/her)
Co-founder, core developer, and steering council member of @napari.org
Also other FOSSy and Science-y things.
I prefer Mastodon (profile at https://fosstodon.org/@jni) but begrudgingly checking things out here. 😂
Microscopist and Bio-Image Analyst for the BioImaging and Optics Platform https://biop.epfl.ch/ at EPFL
https://nicolas.chiaruttini.org
Since 1989, The Institute for Bird Populations has enabled science-based conservation of species and habitats by studying the abundance, demography, and ecology of birds and other wildlife.
Predoc @savitski_lab @EMBL | Computational Biology @saezlab | Interested in functional proteomics and systems biology 🧫🧬
PhD student @ EPFL developing computational methods for microscopy images. Prev Data Science @ UPC
The international network of imaging facilities and communities - empowering imaging scientists across the globe.
The bluesky account is operated by http://quarep.org/impressum. Privacy policy: https://quarep.org/about/privacy-policy/
Astronomy lover, PhD on retinal neural cells stuff, microscopy facility staff. Can you see the connection?
Building CellWalk • Apple Design Awards Finalist
computer graphics • phd • biology • visionOS
User of microscopes. Interested in organelles and how they move. Husband, dad, intermediate filament apologist, and postdoc in the JLS lab at HHMI Janelia Research Campus.
#Duke #UCSD Alum; Recovering Academic; Research IT
@jacksonlab.bsky.social; core dev @napari.fosstodon.org.ap.brid.gy
n-dimensional image viewer for Python
Medical faculty of the University of Heidelberg, Aulehla group @EMBL Heidelberg 🎏
✨Me gusta ver cosas pequeñas de colores y jugar con píxeles :)
🔬BioImage Analyst - Postdoctoral Associate @bethcimini.bsky.social
🍜Like making food
🐲Beginner DM
(He/Him)