FEBS Press
Cells rely on regulated proteostasis mechanisms to keep their internal compartments functioning properly. When these mechanisms fail, damaged proteins accumulate, disrupting organelles, such as the n....
Our new review on proteostasis of organelles in aging & disease is now online in The FEBS Journal!
We discuss recent insights on how #proteostasis safeguards organelle integrity and function across the cell in #aging and disease
doi.org/10.1111/febs...
05.02.2026 08:34 — 👍 15 🔁 4 💬 1 📌 1
YouTube video by David Fay
David's AlphaFold WorkShop 2026
Greetings! I decided to make a YouTube video of my AlphaFold workshop that I've given a few times in the past year. Caveats aside, people seem to find this useful for thinking about how to model protein interactions and how to interpret various AF outputs 1/2
www.youtube.com/watch?v=u63o...
02.01.2026 23:03 — 👍 102 🔁 32 💬 2 📌 1

We are thrilled to share our new pre-print on the molecular anatomy of nuclear proteasome in human sperm cells! #In-cellcryo-ET + #SPA + #LM! @piotrkolata.bsky.social @a-dsantos.bsky.social, @tomdendooven.bsky.social!! doi.org/10.64898/202...
19.12.2025 08:14 — 👍 87 🔁 21 💬 3 📌 1

UBA6 specificity for ubiquitin E2 conjugating enzymes reveals a priority mechanism of BIRC6
Nature Structural & Molecular Biology - Riechmann et al. uncover structural features governing ubiquitin transfer from ubiquitin-activating E1 enzymes UBA1 and UBA6 to specific E2...
Excited to share our work into ubiquitin E1-E2 specificity mechanisms. CryoEM visualising #ubiquitin transfer,biochemistry,evolution @labhofmann.bsky.social & tissue expression analyses @psarkies.bsky.social Big thanks to the team,reviewers & handling editor @dimitristypas.bsky.social rdcu.be/eTRdR
09.12.2025 13:21 — 👍 34 🔁 13 💬 4 📌 2

We’ve made some new tools to manipulate N-recognins. Check them out in our preprint.
www.biorxiv.org/content/10.1...
29.11.2025 03:24 — 👍 18 🔁 13 💬 0 📌 0

ZAK activation at the collided ribosome - Nature
The kinase ZAK is activated at collided ribosomes to mediate the ribotoxic stress response.
Check out our latest work on how collided ribosomes activate the MAP3K ZAK! 💫
www.nature.com/articles/s41...
A fun collaboration with @beckmannlab.bsky.social @doubleshuang.bsky.social
24.11.2025 21:12 — 👍 44 🔁 23 💬 0 📌 1

MISO: microfluidic protein isolation enables single-particle cryo-EM structure determination from a single cell colony.
Or from a single dish of HEK cell culture in the case of two membrane proteins.
Out in Nature Methods now! lnkd.in/gpyBSceg
Wonderful collaboration with the Efremov lab.
14.11.2025 18:38 — 👍 94 🔁 44 💬 3 📌 5
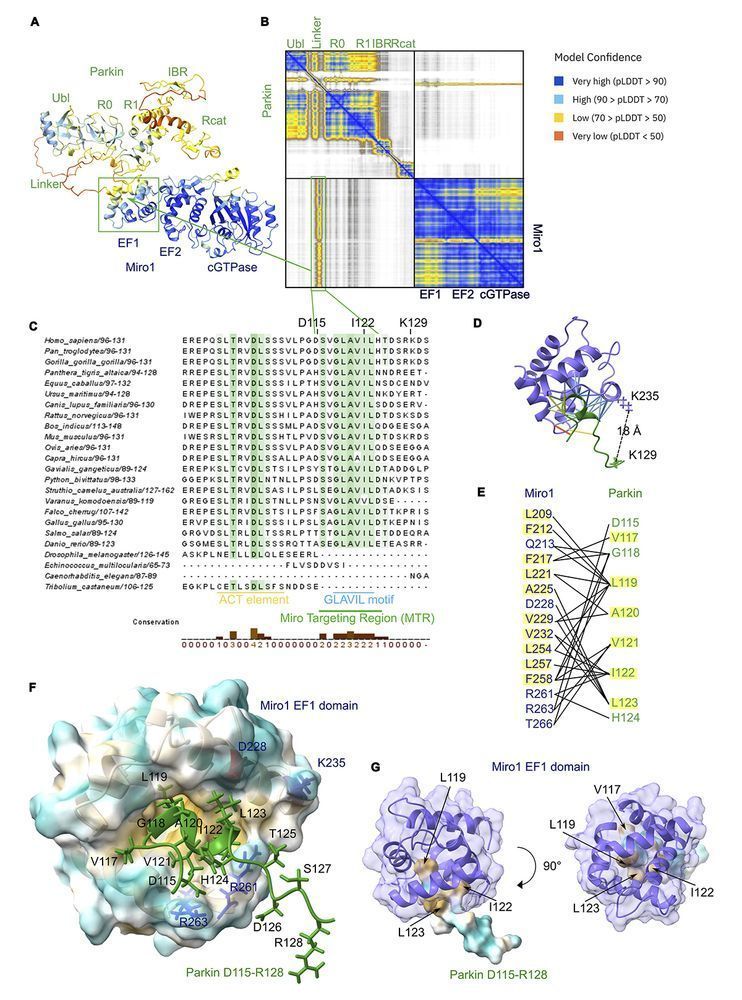
The E3 ligase Parkin ubiquitinates many proteins. Koszela, Walden (@hw1o9.bsky.social) et al. @uofglasgow.bsky.social identify and biochemically validate a direct Parkin interaction with a substrate, Miro1. rupress.org/jcb/article/...
27.06.2025 20:10 — 👍 36 🔁 13 💬 0 📌 3
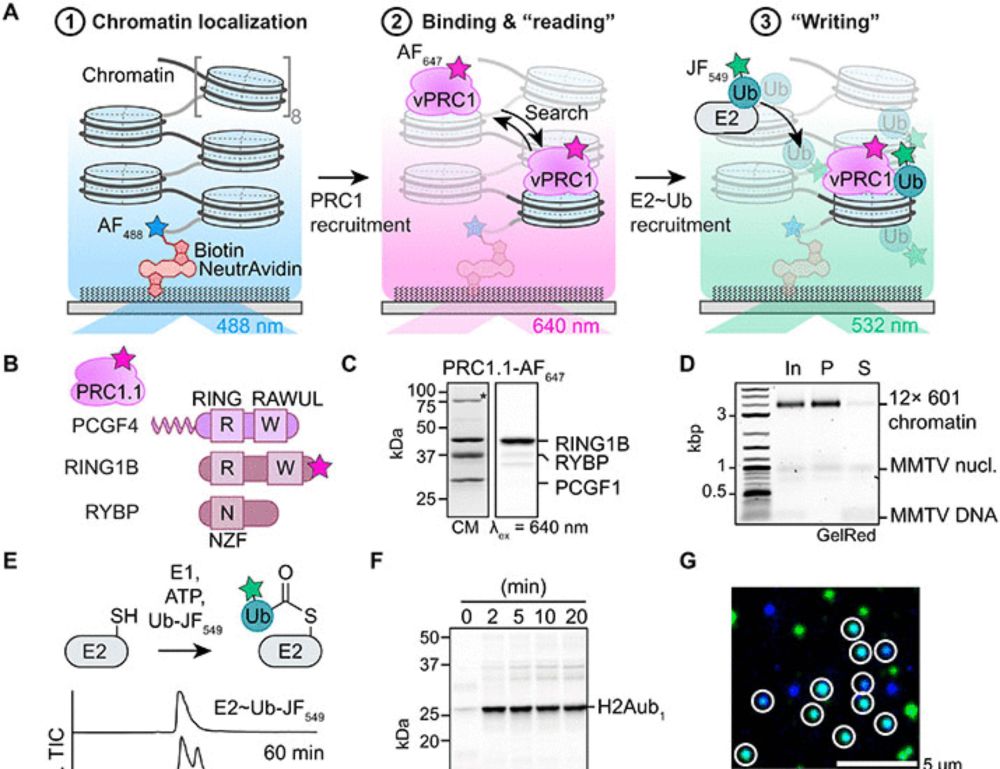
Single-molecule analysis reveals the mechanism of chromatin ubiquitylation by variant PRC1 complexes
Single-molecule experiments show that active conformation formation controls chromatin ubiquitylation kinetics by variant PRC1.
Our new study of chromatin ubiquitylation by variant PRC1 on the single-molecule scale: We visualize directly how vPRC1 ubiquitylates neighboring nucleosomes during a single binding event, showing a potential mechanism how H2Aub domains are established.
www.science.org/doi/10.1126/...
21.05.2025 18:59 — 👍 74 🔁 29 💬 2 📌 0
Molecular basis of SIFI activity in the integrated stress response - Nature
Nature - Molecular basis of SIFI activity in the integrated stress response
Excited that our work revealing the structure and function of the monster stress response silencing factor, the E3 ligase SIFI, is now out! Congratulations to the most amazing team of Zhi Yang, Diane Haakonsen, Michael Heider and Sam Witus!
www.nature.com/articles/s41...
07.05.2025 10:27 — 👍 45 🔁 14 💬 1 📌 2
You want to start tomography? Solve structures inside cells? Reach Nyquist 😳 ? @phaips.vd.st and I have a website for you! tomoguide.github.io
You'll find a tutorial on how to reconstruct tomograms, pick particles and do subtomogram averaging, using different software!
Hope it will be useful !
06.05.2025 16:42 — 👍 125 🔁 37 💬 3 📌 0

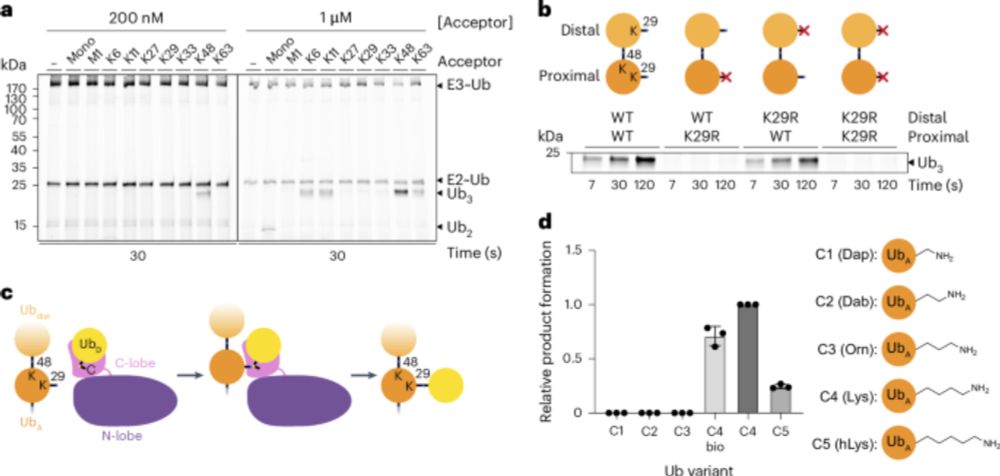
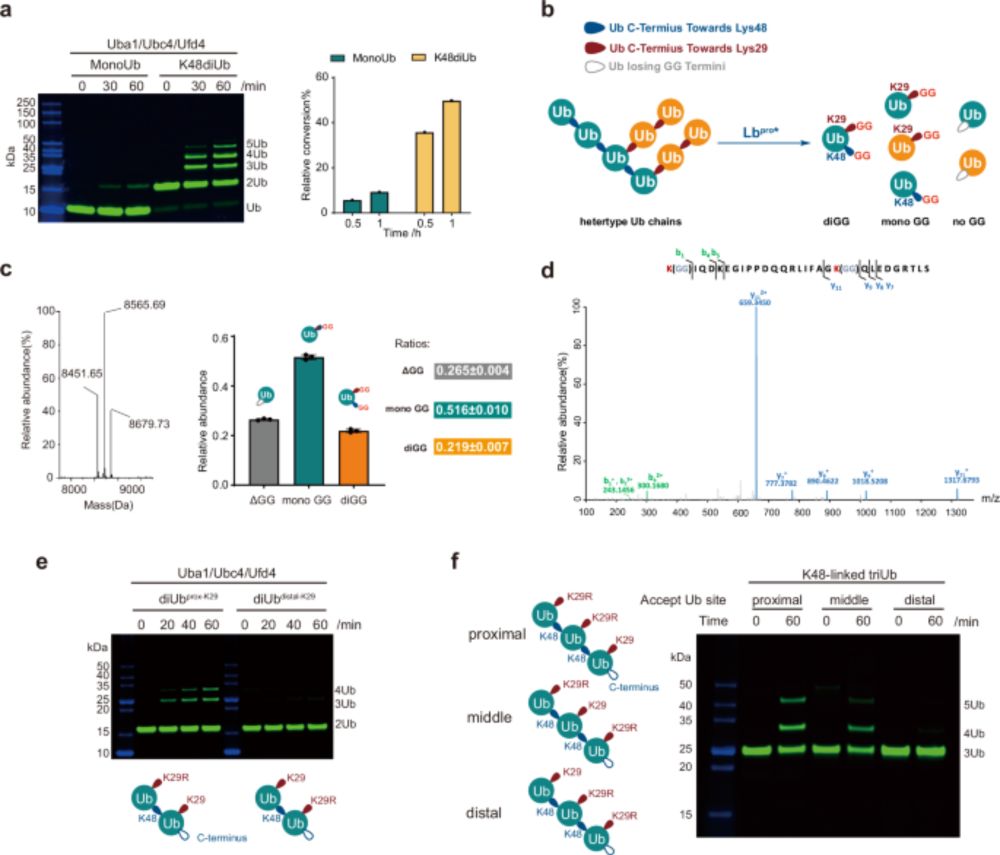
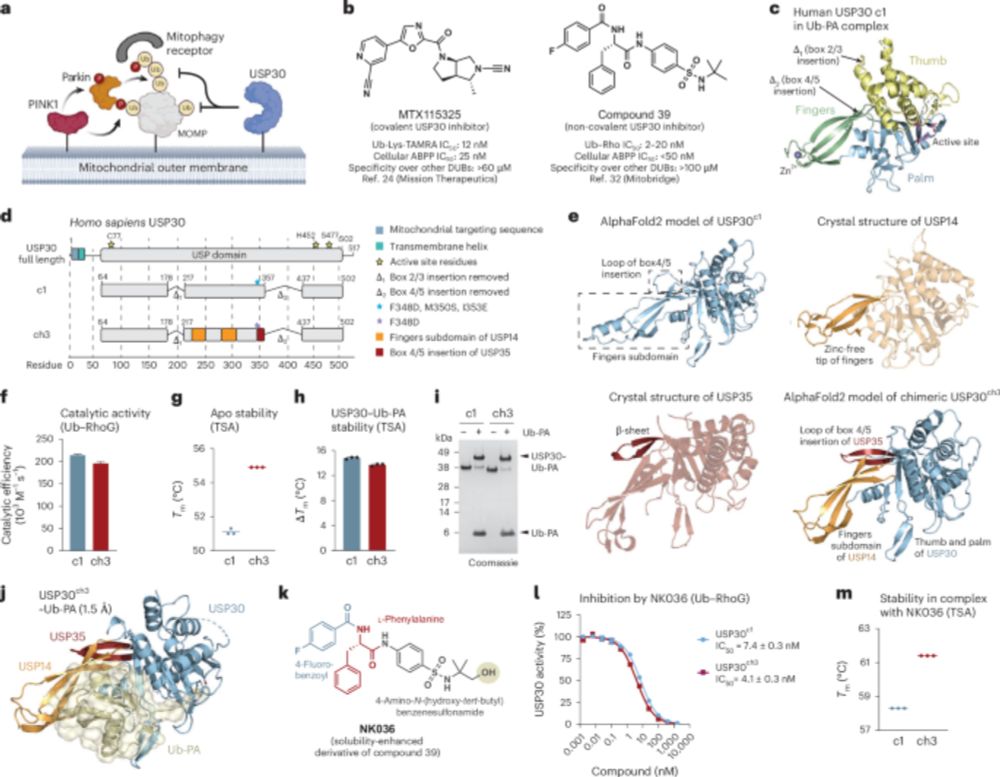
Happy to see our review out in ChemBioChem @chemistryeurope.bsky.social in which we describe the tools and strategies for deciphering linkage type-specific #ubiquitin signalling: doi.org/10.1002/cbic.... Thanks to Julian for spearheading this, and to Camilla and Dominik for important contributions.
22.04.2025 11:31 — 👍 31 🔁 9 💬 1 📌 0

Retromer promotes the lysosomal turnover of mtDNA
Lysosomal uptake of mtDNA controls mitochondrial quality.
Excited to share our latest paper in Science Advances! We finally dissected a new pathway for mitochondrial quality control upon mtDNA replication stress! Let’s start a short thread! 🧵
www.science.org/doi/10.1126/...
04.04.2025 18:42 — 👍 78 🔁 28 💬 6 📌 3
PhD student in Maddika Subbareddy Lab @bric-cdfd.bsky.social
Ubiquitin and Nedd8 enthusiast | works on Dynein motor.
My interests: Science, SciComm, Contemporary fiction & Mental Health
muckrack.com/devanshigupta
🇺🇦 Associate professor UCONN Health. NMR, structural biology, ubiquitination, sumoylation, DUBs, sciArt
Research Assistant Professor of Biochemistry and Biophysics | Director, Johnson Foundation Biophysics and Structural Biology Core Facility | Univ of PA | he/him
Fascinated by ubiquitination. Works at the RWTH Aachen University. Tweets are my own.
Research Group Leader at the University of Edinburgh. Using genome editing and targeted protein degradation to study mechanisms underlying human disease and therapy.
Using molecular biophysics, biochemistry, and cell biology to understand the intersection of protein quality control mechanisms and neurodegenerative disorders (ALS, FTD); IDPs, Ub biology, UBQLNs
Postdoc in Komander lab @wehi-research.bsky.social | Protein Structure-Function | Ubiquitin
rashmiagrata.com
Group Leader, Induced Proximity Therapeutics, Centre for Protein Degradation, Institute of Cancer Research, London
Scientific illustration, Figure Design, Logo Creation, and Web Design
We transform discoveries into artistic masterpieces #SciArt
✉️ contact@drawimpacts.com
Professor for Medical Biochemistry at Saarland University, Germany. My lab studies the biogenesis and function of lipid droplets
A chembio group @Temple Chemistry & Fox Chase Cancer Center. We work on small molecules, peptides & protein probes, expanding toolkits for proteomics & cellular imaging, with the goal of deciphering cell biology. Find more @ https://rosswang.weebly.com
Assistant Prof at UWaterloo in Biology. Study autophagy & protein lipidation (S-acylation & N-myristoylation) in ALS & Huntington disease. Expect science, food, some cdnpoli, & dog pics
https://neurdyphagylab.squarespace.com/
Prof at University of Michigan, Cell & Developmental Biology. Microtubules, Motor Proteins, Microscopy.
Biochemist and Cell Biologist, Professor @unicologne.bsky.social, Director Center for Biochemistry @bccologne.bsky.social
PhD student in Hiller lab, University of Basel | studying molecular chaperones and protein folding homeostasis
Toxin-Antitoxin Systems, Molecular Chaperones, group leader CNRS Toulouse
Biophysicist/Statistical physicist, Protein Quality Control (chaperones galore), energy-driven processes in the cell, machine learning/AI for proteins, and origins of life @EPFL, Lausanne.
Opinions are mine.
#SempreForzaToro
PI at Leibniz Institute on Aging, Jena + Professor for Biochemistry at University of Jena, Germany
We study amyloid proteins, molecular chaperones in the context of aging.
Group leader at the Francis Crick institute and head of the Protein Biogenesis lab. Interested in protein folding, ribosomes and molecular chaperones.
Group leader, Charlotte Group for Proteostasis Research | Associate Chair for Research in Biological Sciences at UNC Charlotte | Interested in the role of Hsp70 PTMs in yeast and mammalian cells | www.trumanlab.org