How can we extract unbiased transition rates from ligand-unbinding Random Acceleration MD (RAMD) simulations? Our new paper lays out the theoretical framework behind RAMD. Take a look! doi.org/10.1063/5.03... @benoitroux.bsky.social @yiweiding.bsky.social & Alessia Ghidini #MD #drugdiscovery
20.02.2026 18:35 —
👍 11
🔁 5
💬 0
📌 0

Your protein moves, but when exactly? ⏱️
Meet eRMSF, our new Python tool that tracks fluctuations over time!
Traditional RMSF shows “how much.”
eRMSF shows “when.” 🔥
Preprint 🔗 doi.org/10.26434/che...
GitHub github.com/pablo-arante...
Try it on Colab → colab.research.google.com/github/pablo...
07.10.2025 13:12 —
👍 4
🔁 1
💬 0
📌 0


Really proud of my sister’s sustainable knitwear business. She’s making some bold statement pieces! www.etsy.com/uk/shop/Grac...
16.09.2025 11:06 —
👍 11
🔁 3
💬 1
📌 0

Design of multi-target peptide modulators for protein chaperone networks
Essential chaperones heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) collaborate in oncoprotein folding. Dual inhibition of these chap…
🚀New adventures in #DrugDiscovery: We designed multi-target leads that hit Hsp70, Hsp90 & Cdc37 at once—disrupting chaperone networks that fuel tumor growth. 🧬💥 Great collaboration with Mollapour @medmol.bsky.social and Gestwicki Labs
#CancerResearch #CompChem
shorturl.at/gGk7m
26.08.2025 09:31 —
👍 8
🔁 1
💬 1
📌 0
We are looking for someone to join the group as a postdoc to help us with scaling implicit transfer operators. If you are interested in this, please reach out to me through email. Include CV, with publications and brief motivational statement. RTs appreciated!
27.05.2025 13:23 —
👍 15
🔁 8
💬 1
📌 2

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
08.05.2025 16:06 —
👍 141
🔁 58
💬 4
📌 9
Once again, a great collaboration between the Buchner Lab, the @sattler-lab.bsky.social and the Rosenzweig Lab!
30.04.2025 13:38 —
👍 1
🔁 1
💬 0
📌 0

Google Colab
Run BioEmu in Colab - just click "Runtime → Run all"! Our notebook uses ColabFold to generate MSAs, BioEmu to predict trajectories, and Foldseek to cluster conformations.
Thanks @jjimenezluna.bsky.social for the help!
🌐 colab.research.google.com/github/sokry...
📄 www.biorxiv.org/content/10.1...
29.03.2025 09:50 —
👍 102
🔁 42
💬 1
📌 2
✨✨ It's #glycotime for #HIV Env ✨✨
N-linked glycans modulate flexibility & MPER epitope exposure #glycotime
Huge effort by @shehata92.bsky.social @lcasalino88.bsky.social with cryoET of Env in VLPs by @thevillalab.bsky.social & team 💪
Would love feedback!
www.biorxiv.org/content/10.1...
28.03.2025 04:23 —
👍 144
🔁 31
💬 6
📌 5
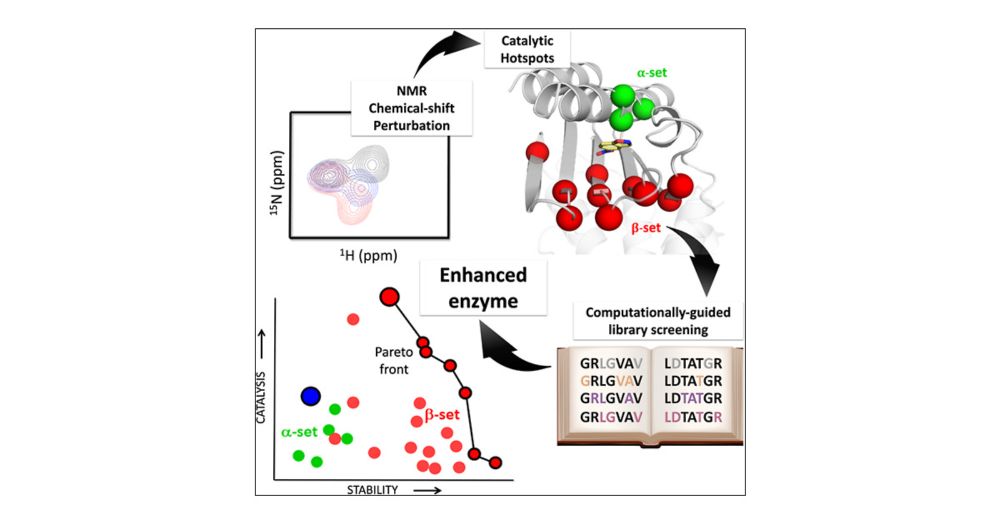
Enzyme Enhancement Through Computational Stability Design Targeting NMR-Determined Catalytic Hotspots
Enzymes are the quintessential green catalysts, but realizing their full potential for biotechnology typically requires improvement of their biomolecular properties. Catalysis enhancement, however, is often accompanied by impaired stability. Here, we show how the interplay between activity and stability in enzyme optimization can be efficiently addressed by coupling two recently proposed methodologies for guiding directed evolution. We first identify catalytic hotspots from chemical shift perturbations induced by transition-state-analogue binding and then use computational/phylogenetic design (FuncLib) to predict stabilizing combinations of mutations at sets of such hotspots. We test this approach on a previously designed de novo Kemp eliminase, which is already highly optimized in terms of both activity and stability. Most tested variants displayed substantially increased denaturation temperatures and purification yields. Notably, our most efficient engineered variant shows a ∼3-fold enhancement in activity (kcat ∼ 1700 s–1, kcat/KM ∼ 4.3 × 105 M–1 s–1) from an already heavily optimized starting variant, resulting in the most proficient proton-abstraction Kemp eliminase designed to date, with a catalytic efficiency on a par with naturally occurring enzymes. Molecular simulations pinpoint the origin of this catalytic enhancement as being due to the progressive elimination of a catalytically inefficient substrate conformation that is present in the original design. Remarkably, interaction network analysis identifies a significant fraction of catalytic hotspots, thus providing a computational tool which we show to be useful even for natural-enzyme engineering. Overall, our work showcases the power of dynamically guided enzyme engineering as a design principle for obtaining novel biocatalysts with tailored physicochemical properties, toward even anthropogenic reactions.
Our latest paper "Enzyme Enhancement Through Computational Stability Design Targeting NMR-Determined Catalytic Hotspots" now out in JACS @jacs.acspublications.org. Collaboration with Jose M. Sanchez-Ruiz in Granada, among others. pubs.acs.org/doi/full/10....
20.03.2025 02:00 —
👍 41
🔁 13
💬 4
📌 0
This sounds great. Also, what a lot of questions visualizations like this raise. Don't you think, @bnerlich.bsky.social?
20.03.2025 18:09 —
👍 45
🔁 16
💬 5
📌 0

Point mutations of the mitochondrial chaperone TRAP1 affect its functions and pro-neoplastic activity
Cell Death & Disease - Point mutations of the mitochondrial chaperone TRAP1 affect its functions and pro-neoplastic activity
Read our latest research with the Rasola Lab and @elenapapaleo.bsky.social on how mutations on the mitochondrial protein TRAP1 impact its function and cancer related activities, published with
@springernature on Cell Death and Disease in rdcu.be/edidL
12.03.2025 16:35 —
👍 7
🔁 1
💬 0
📌 0
Redirecting
Happy to share our latest work "Large-scale energy decomposition for the analysis of protein stability". We use simple simulations to predict the impact of mutations on stability of more than 250 mutants in different folds. doi.org/10.1016/j.cs...
#CompChem #CompBiol #AI #Biotechnology
06.02.2025 07:42 —
👍 4
🔁 0
💬 0
📌 0
Postdoc position in Structural Biology for Drug Discovery
Fare clic sul collegamento fornito per visualizzare la descrizione completa dell'offerta di lavoro.
Please check it out - postdoc is available in Struct Biol for Drug Discovery in Cancer in my group at IIT.
Great project with potential for impactful results, papers and more ...
iit.taleo.net/careersectio...
04.02.2025 12:16 —
👍 6
🔁 6
💬 0
📌 0
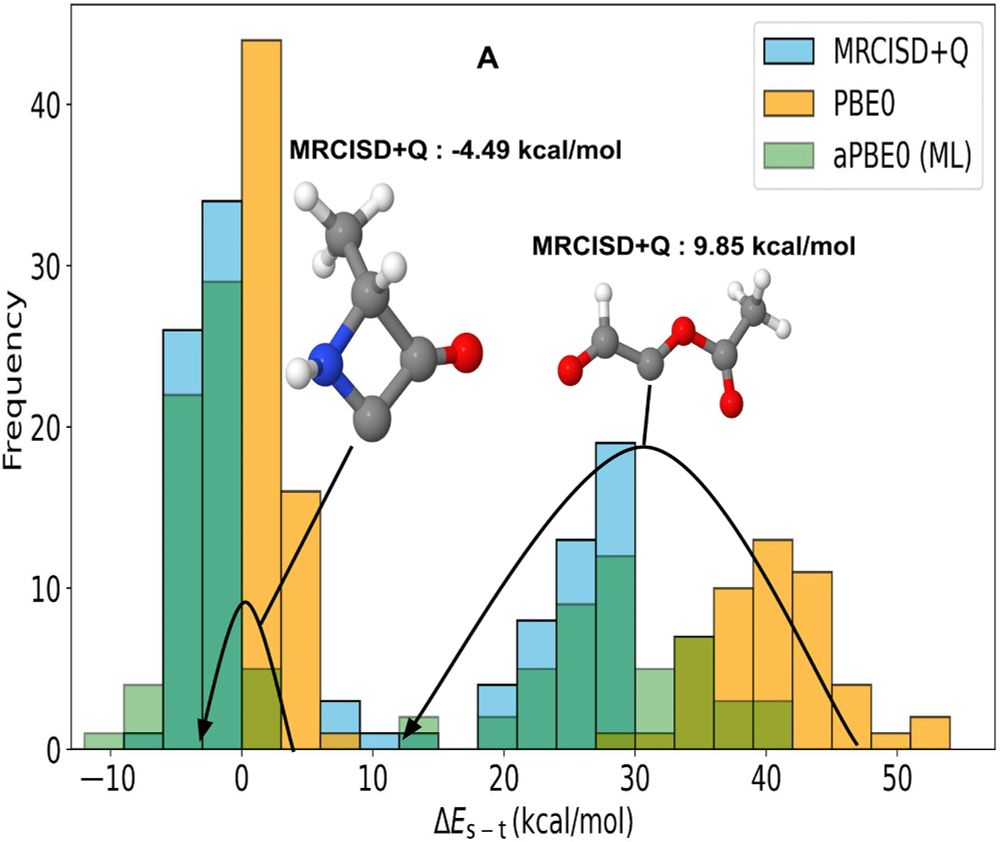
Pumped about #MachineLearning the adaptive exact exchange admixture in hybrid #DFT approximations: It can even cure the infamous spin-gap problem (see below). Just out in @ScienceAdvances with D Khan, A Price, B Huang and M Ach! @uoft.bsky.social #CompChem www.science.org/doi/10.1126/...
03.02.2025 17:23 —
👍 13
🔁 5
💬 0
📌 1

Impact of Ligand-Induced Oligomer Dissociation on Enzyme Diffusion, Directly Observed at the Single-Molecule Level
The existence of the phenomenon of enhanced enzyme diffusion (EED) has been a topic of debate in recent literature. One proposed mechanism to explain the origin of EED is oligomeric enzyme dissociation. We used mass photometry (MP), a label-free single-molecule technique, to investigate the dependence of the oligomeric states of several enzymes on their ligands. The studied enzymes of interest are catalase, aldolase, alkaline phosphatase, and vanillyl-alcohol oxidase (VAO). We compared the ratios of oligomeric states in the presence and absence of the substrate as well as different substrate and inhibitor concentrations. Catalase and aldolase were found to dissociate into smaller oligomers in the presence of their substrates, independently of inhibition, while for alkaline phosphatase and VAO, different behaviors were observed. Thus, we have identified a possible mechanism which explains the previously observed diffusion enhancement in vitro. This enhancement may occur due to the dissociation of oligomers through ligand binding.
Proud to announce Yulia's paper in Nano Letters. We were planning to measure enzyme diffusion rates under different conditions using label-free single-molecule methods, and accidentally uncovered an interesting effect: pubs.acs.org/doi/10.1021/... 🧪
30.01.2025 16:21 —
👍 34
🔁 8
💬 2
📌 2
Don't miss your chance to join this excellent @bioexcelcoe.bsky.social seminar series: Next option tomorrow! 🧐
27.01.2025 12:20 —
👍 3
🔁 3
💬 0
📌 0

The neglected ‘laws’ of chemistry – and why they matter | Aeon Essays
Often dismissed as the poor cousin of the sciences, chemistry has revealed natural laws that illuminate our Universe
'Everyone claimed some form of superiority. Physicists would say that chemistry is just applied physics; chemists would say that biology is just applied chemistry; biologists would claim that life’s complexity cannot be captured solely by analysing atoms and molecules.'
aeon.co/essays/the-n...
25.01.2025 09:09 —
👍 6
🔁 3
💬 1
📌 2
























