Fast execution - 5 mins runtime for 200 samples using combined segmentation (versus 40 mins runtime for established algorithms conumee and conumee 2)
#DNAmethylation #EPICv2 #bioinformatics #braintumor #neurooncology #neuropathology #ukeHamburg #zmnh #diagnostics
24.09.2025 19:05 —
👍 1
🔁 0
💬 0
📌 0
Broad applicability - single sample or combined sample analyses, all array types (450K, EPIC, EPICv2, mouse), integration of conumee and conumee 2
Flexible visualisation - Intensity Plot and Frequency Plot
High sensitivity. - Up to 96% for all array types for focal chromosomal aberrations
24.09.2025 19:05 —
👍 0
🔁 0
💬 1
📌 0
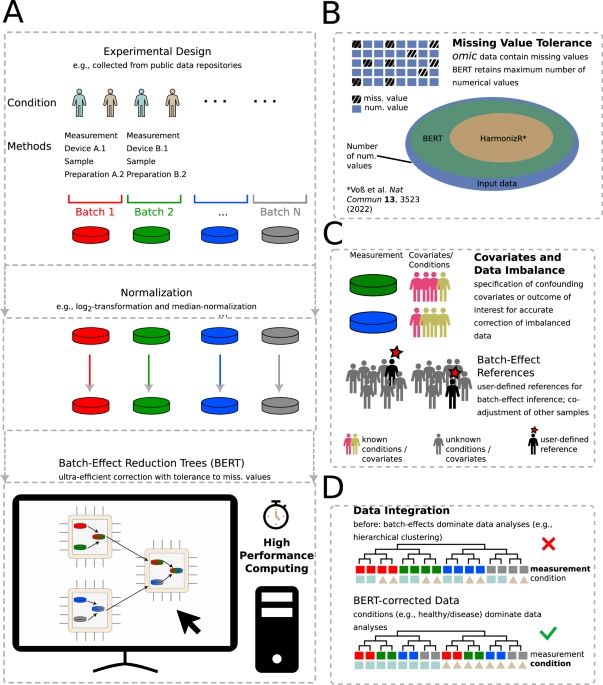
The Cumulative CNV (CCNV) R package combines established segmentation methods and a newly implemented algorithm for thorough and fast CNV analysis at unprecedented accessibility enabling the analyses of large case series.
Advantages:
Easy to use - Only one package and line of code
24.09.2025 19:05 —
👍 0
🔁 0
💬 1
📌 0
Copy number variation (CNV) patterns can be inferred from global DNA methylation data, which are commonly analyzed in routine brain tumor diagnostics.
CNV analyses depict alterations of DNA quantities across chromosomes that are crucial for tumor diagnostics and classification.
24.09.2025 19:05 —
👍 0
🔁 0
💬 1
📌 0
8. BERT was thoroughly tested emphasizing it's broad scope (tested on large-scale data integration tasks with up to 5000 datasets from simulated and experimental data of different quantification techniques and omic types (proteomics, transcriptomics, metabolomics) as well as other datatypes)
02.08.2025 15:56 —
👍 0
🔁 0
💬 0
📌 0
6. BERT is super fast (leverages multi-core and distributed-memory systems for up to 11 × runtime improvement)
7. BERT is available on Bioconductor
02.08.2025 15:56 —
👍 0
🔁 0
💬 1
📌 0
3. BERT retains data in incomplete datasets upon integration
4. BERT can integrate datasets with different compositions and conditions (integration of unbalanced datasets using covariates)
5. BERT can integrate datasets with unknown conditions (using references)
02.08.2025 15:56 —
👍 0
🔁 0
💬 1
📌 0
BERTs features:
1. BERT can integrate molecular data (Omic data) or other data types (e.g. clinical annotations)
2. BERT tolerates missing data of different types in datasets (e.g. missing at random, missing not at random)
02.08.2025 15:56 —
👍 0
🔁 0
💬 1
📌 0
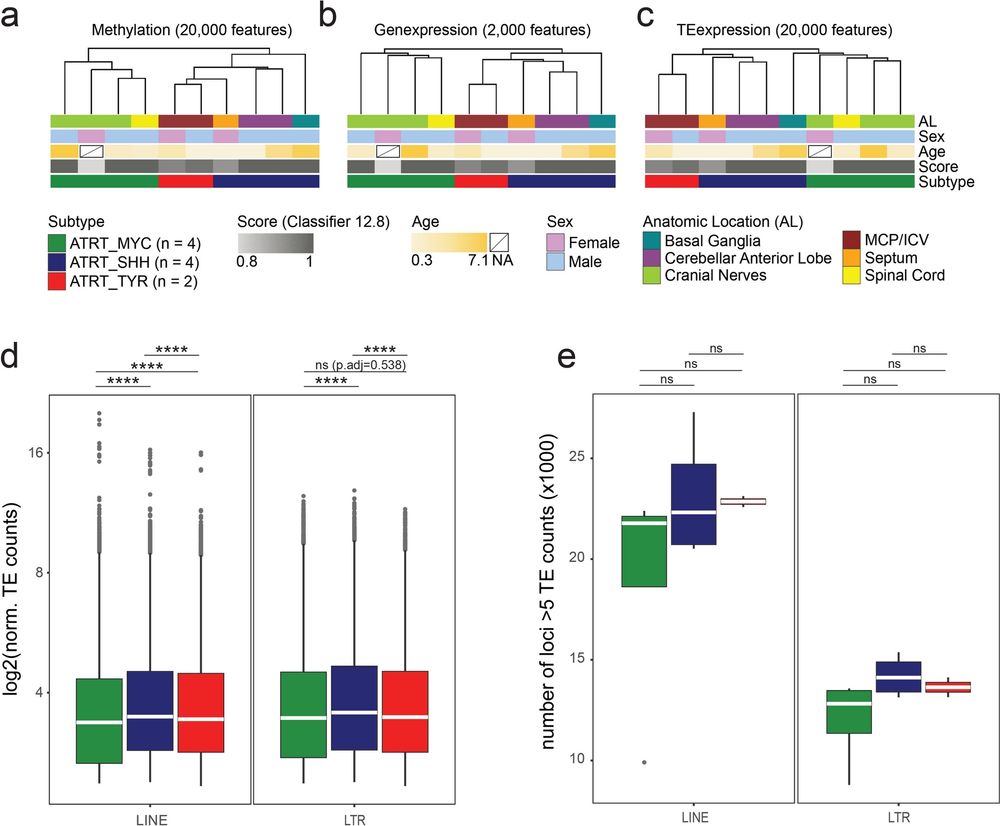
Including TE transcription profiles into the molecular characterization of ATRTs might reveal new tumor vulnerabilities leading to novel therapeutic interventions, such as immunotherapy.
23.07.2025 19:58 —
👍 0
🔁 0
💬 0
📌 0
--> Differentially transcribed TEs in primary samples are partly reflected in established ATRT-SHH and -MYC cell line models.
23.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0
--> ATRT-MYC showed broadly reduced transcript levels of LINE1 and ERVL-MaLR subfamilies and displayed significantly less LTR and LINE1 loci with bidirectional promoter activity.
23.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0
--> We investigate the transcriptional profiles of 1.9M LINE1 and LTR elements that differ across ATRT subtypes in primary human samples.
23.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0
Atypical teratoid rhabdoid tumors (ATRTs) are very aggressive CNS tumors mainly affecting infants.
Transcriptional activity of transposable elements (TEs), like LINE1s and LTRs, is tightly linked with human cancers as a direct consequence of lifting epigenetic repression over TEs.
23.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0
Many thanks to the @dfg.de for funding this research within the #emmynoether programme!
22.07.2025 21:04 —
👍 0
🔁 0
💬 0
📌 0
We provide molecular insights into these rare and severe brain diseases and reveal novel implications of the oncogenic factor LIN28A in extracellular matrix integrity
22.07.2025 21:04 —
👍 0
🔁 0
💬 1
📌 0
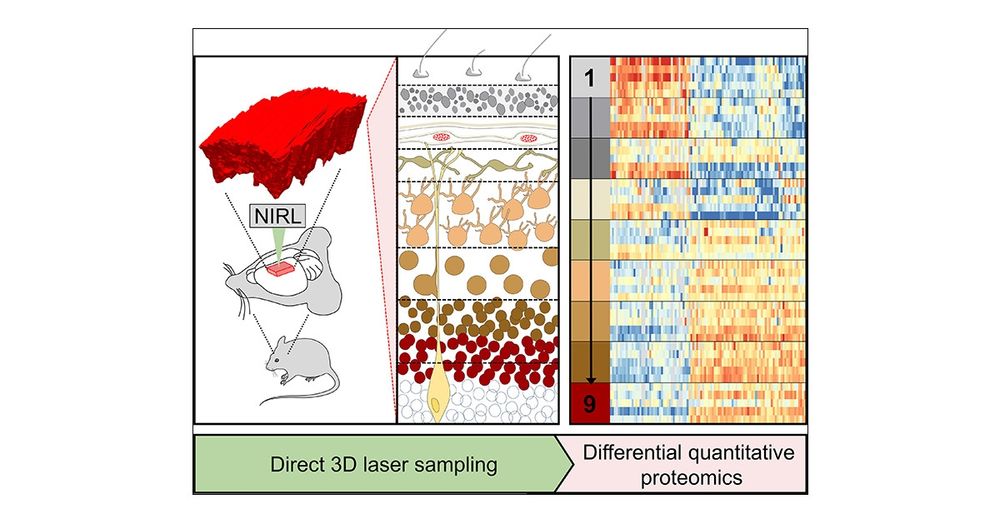
Alterations of brain layer morphology in these models were mapped to spatial proteome patterns that were acquired with the Nanosecond-infrared laser system
22.07.2025 21:04 —
👍 0
🔁 0
💬 1
📌 0
•
In contrast sole stabilisation of CTNNB1 resulted in a distinct developmental brain phenotype similar to Lissencephaly Type 1.
22.07.2025 21:04 —
👍 1
🔁 0
💬 1
📌 0
In a nutshell:
•
Co-activation of LIN28A and stabilised CTNNB1 in vivo led to pial disruption and neuronal overmigration resembling a servere human developmental disorder of the brain which is called the Cobblestone Lissencephaly Type 2.
22.07.2025 21:04 —
👍 0
🔁 0
💬 1
📌 0
Jelenas work on #spatialproteomics of the developing brain has now been published in Molecular md Cellular Proteomics (www.mcponline.org/article/S153... )
#neurooncology #neurodevelopment #proteomics #Neuropathology #zmnh #ukeHamburg
22.07.2025 21:04 —
👍 2
🔁 1
💬 1
📌 0
Finally, hypomethylation at the FGFR3 locus, together with increased FGFR3 protein expression, was observed in 97% of cases, highlighting FGFR3 as a potential future treatment target.
13.06.2025 21:08 —
👍 0
🔁 0
💬 0
📌 0
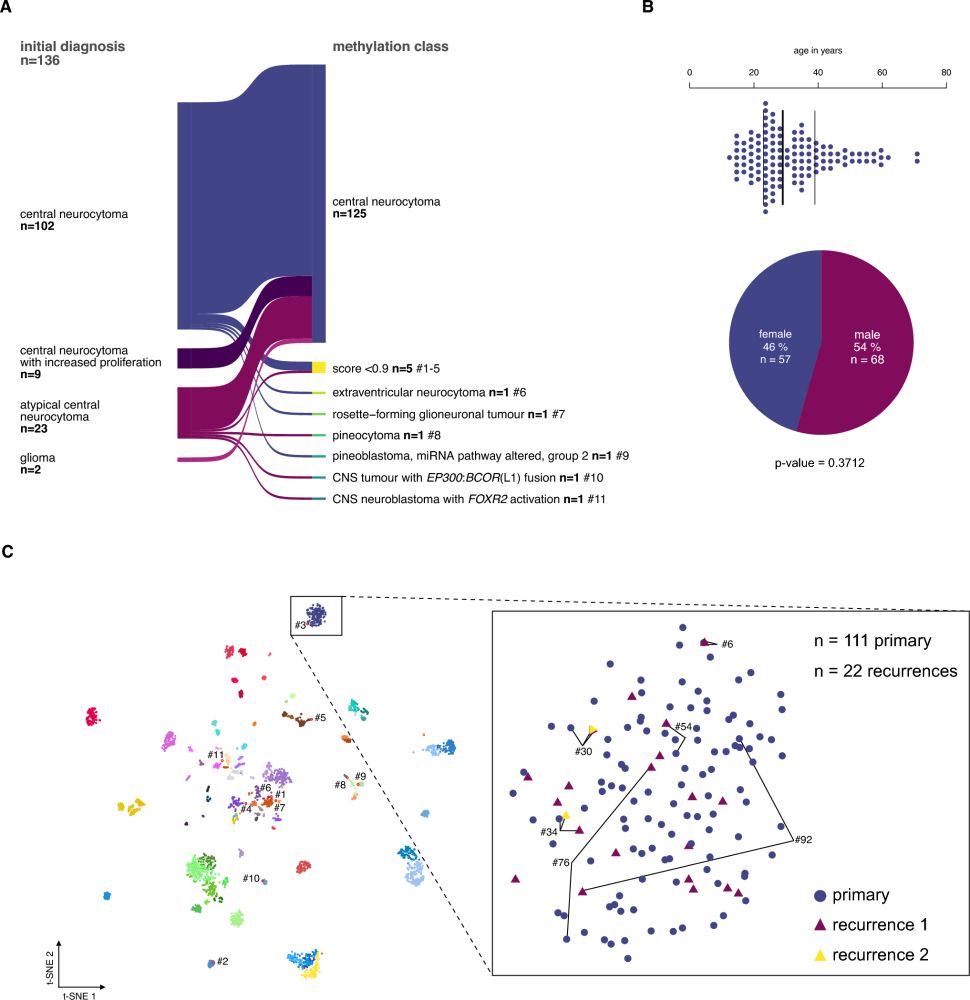
DNA methylation profiles were rather homogenous but global DNA hypomethylation was a feature of tumors associated with higher recurrence risk.
13.06.2025 21:08 —
👍 0
🔁 0
💬 1
📌 0
Of note, classic histopathological criteria such as atypia and the cell proliferation index were not reproducible across neuropathologists.
Instead, patient age and treatment strategy were key factors associated with survival outcomes in the cohort.
13.06.2025 21:08 —
👍 0
🔁 0
💬 1
📌 0
Antonia Gocke representing our group at #SNOPeds2025 !
She shows our data on LIN28A in atypical teratoid rhabdoid tumors and proteome data on a large cohort of #ependymoma. Fantastic job Antonia!
#atrt #braintumor #pediatriccancer #kinderkrebs #proteomics #neurooncology #Neuropathology #zmnh
17.05.2025 19:54 —
👍 2
🔁 0
💬 0
📌 0
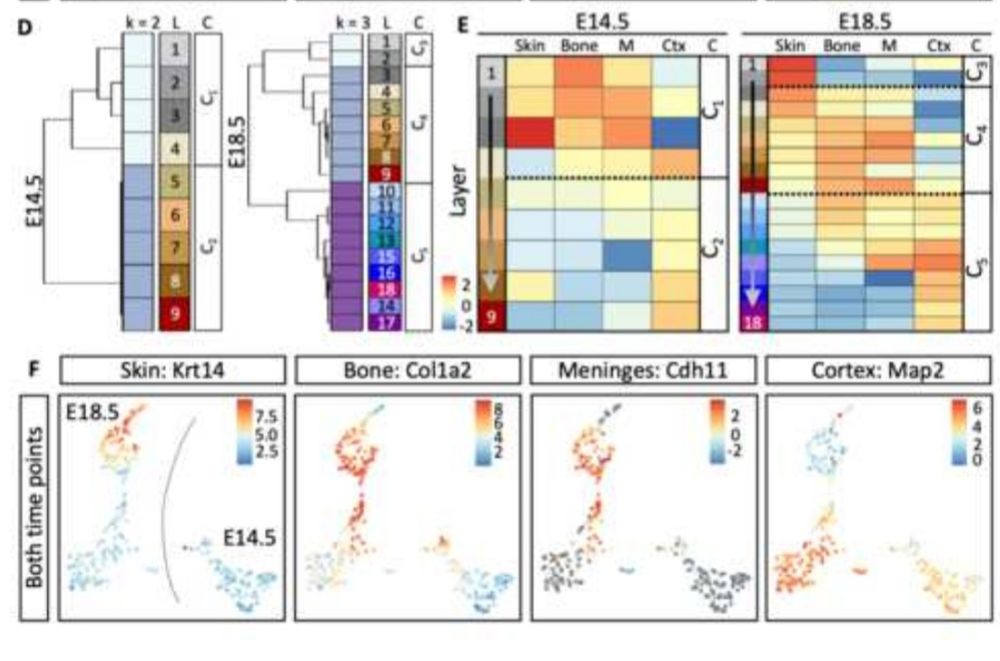
The developing brain cortex is a complex structure relying on a specific spatiotemporal orchestration of signalling molecules and pathways.
To grasp this complexity Jelena Navolic established spatial proteome analyses of the brain applying Nanosecond Infrared Laser Ablation (NIRL).
27.02.2025 19:32 —
👍 0
🔁 0
💬 1
📌 0


Dear Jelena, now you successfully defended your PhD ‐ congratulations! We are very happy to have you in the team!
#proteomics #etmr #spatialproteomics #zmnh #ukeHamburg #neurooncology #neurodevelopment #neuropathology
27.02.2025 19:32 —
👍 3
🔁 0
💬 1
📌 0