Looking forward to the Grand Opening of the Northern Eye Imaging facility on 5th November at the Frederick Douglass Centre, Newcastle. Register now nusbf.ncl.ac.uk/ccpem-tfs-ne... for the latest #structuralbiology and #cryoem in the region, to see the new Tundra cryo-TEM and lots more!
21.10.2025 11:25 — 👍 6 🔁 5 💬 0 📌 1
Rescaling FSC curves pubmed.ncbi.nlm.nih.gov/41059947/ #cryoEM
10.10.2025 01:15 — 👍 4 🔁 2 💬 0 📌 0
Below (by @landerlab.bsky.social) is the 2nd #cryosparc recipe on @biorxivpreprint.bsky.social in a month. I remain worried about closed-source software affecting progress in #cryoEM. We should develop new algorithms instead!
#OpenSoftwareAcceleratesScience
www.biorxiv.org/content/10.1...
05.10.2025 08:20 — 👍 16 🔁 5 💬 3 📌 0
Improved cryo-EM reconstruction of sub-50 kDa complexes using 2D template matching www.biorxiv.org/content/10.1101/2025.09.11.675606v1 #cryoEM
17.09.2025 08:49 — 👍 20 🔁 11 💬 0 📌 1
Confidence-guided cryo-EM map optimisation with LocScale-2.0 www.biorxiv.org/content/10.1101/2025.09.11.674726v1 #cryoEM
15.09.2025 07:11 — 👍 13 🔁 6 💬 0 📌 0
This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
13.09.2025 00:04 — 👍 262 🔁 82 💬 19 📌 14
Magellon has taken off to explore your cryo-EM data! Now out in @iucrj.iucr.org doi.org/10.1107/S205... Congrats to all involved!
05.09.2025 14:48 — 👍 11 🔁 3 💬 0 📌 0

At Darwin College:
Richard Henderson
Jacqueline Milne
Sriram Subramanian
Nigel Unwin
David Barford
Da-neng Wang
John Rubinstein

John ring's the LMB seminar bell to call the audience back in after a coffee break.
Returned from a wonderful symposium in honour of the 50th anniversary of Richard Henderson's and Nigel Unwin's 1975 paper on the structure of bacteriorhodopsin - the paper that started the field of membrane protein structural biology.
Also fulfilled my dream of ringing the LMB seminar bell.
12.07.2025 12:10 — 👍 60 🔁 7 💬 3 📌 0
Ever wonder why SGD, which works so well for ab initio cryo-EM, never seems to get to high resolution on its own? Well, we have the answer: conditioning!
24.06.2025 21:31 — 👍 15 🔁 11 💬 1 📌 0
Standards for MicroED pubmed.ncbi.nlm.nih.gov/40539937/ #cryoEM
21.06.2025 18:41 — 👍 1 🔁 2 💬 0 📌 0
Alves, Andrews, McBean, Wells, El-Gomati, Burkhalter, Schulze-Briese, Zambon, McMullan, Henderson, Russo, Jones: The Dublin Lens: A Cc=1.0 mm Objective Lens Intended for CryoEM at 100 keV https://arxiv.org/abs/2505.11458 https://arxiv.org/pdf/2505.11458 https://arxiv.org/html/2505.11458
19.05.2025 06:14 — 👍 3 🔁 3 💬 1 📌 0
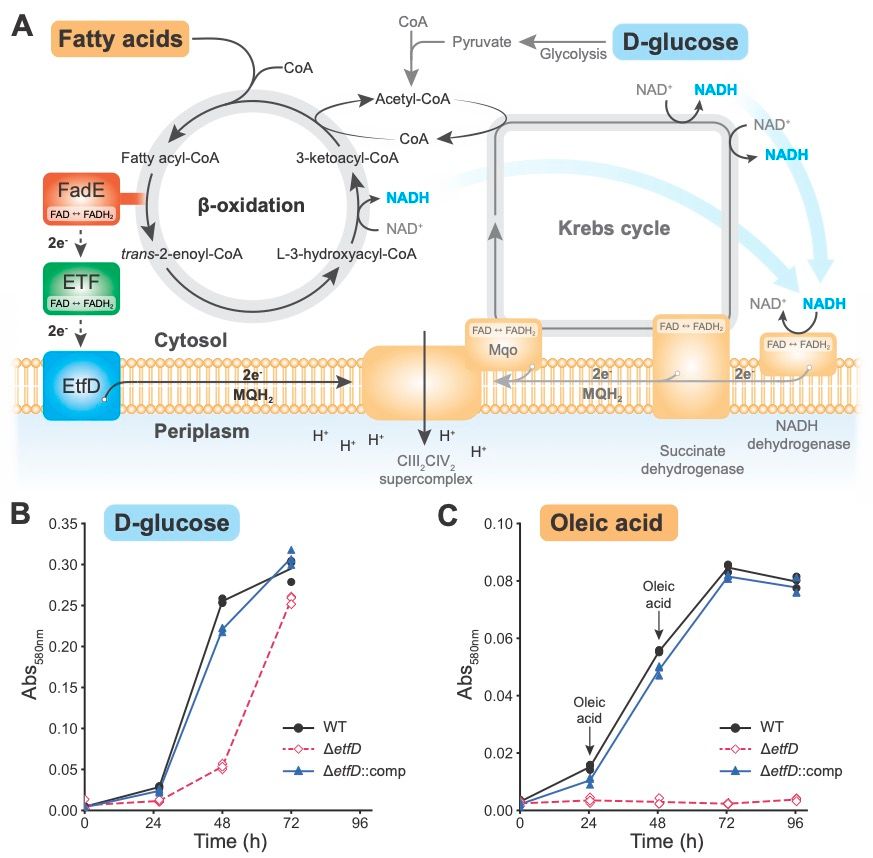
A) Diagram showing how EtfD transfers electrons from beta oxidation to the menaquinol pool in the membrane, linking beta oxidation to oxidative phosphorylation in mycobacteria.
B) A growth curve showing the Mycobacterium smegmatis lacking EtfD can grow when glucose is provided as the carbon source, but has a slight growth delay. Complementing the ∆etfD strain with M. tuberculosis on a plasmid rescues the defect.
C) A growth curve showing that ∆etfD M. smegmatis cannot grow on medium where the fatty acid oleic acid is the only carbon source. A plasmid with with M. tuberculosis EtfD rescues the defect.
EtfD links beta oxidation to OxPhos in mycobacteria - recent work suggests that targeting EtfD could shorten #tubeculosis treatment.
But how does EtfD work? And how can you assay its activity?
In his final PhD manuscript @courbongautier.bsky.social provides answers!
www.biorxiv.org/content/10.1...
16.05.2025 14:30 — 👍 42 🔁 16 💬 1 📌 0
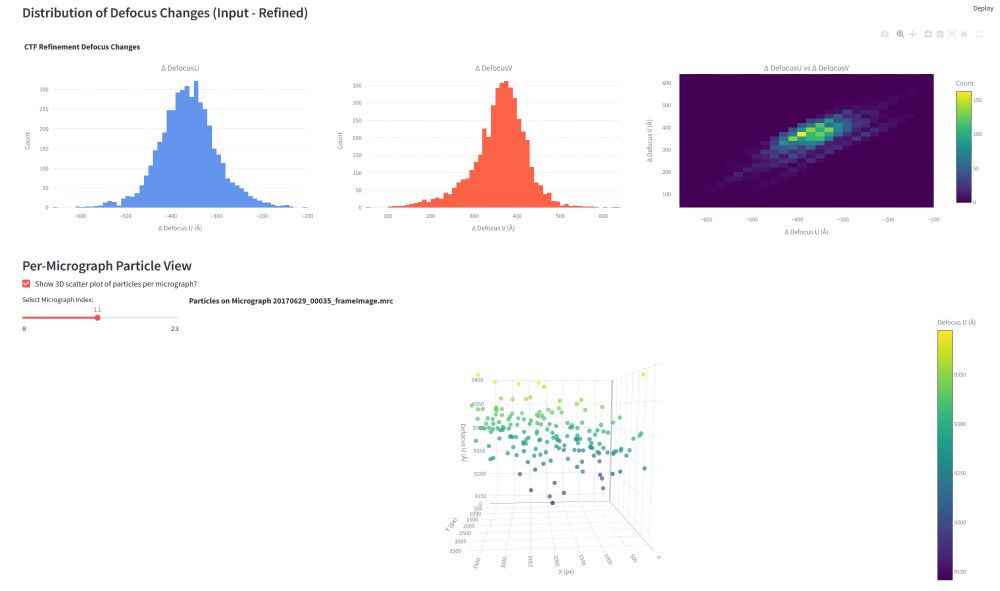
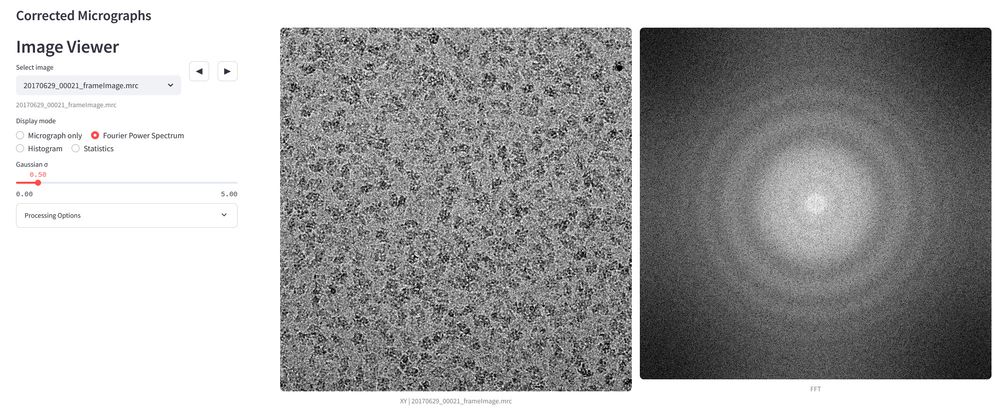
1/5: Over the last few months, I have been working on an update for the #FollowRelionGracefully dashboard, which offers improved real-time job previews for #cryoEM #Relion jobs.
27.04.2025 20:49 — 👍 50 🔁 20 💬 2 📌 2
Close to recreating it in ChimeraX:
1. Install the python file: github.com/smsaladi/chi...
2. rainbow palette magma
3. col modify #1 blackness - 40
4. col modify #1 whiteness - 40
5. col modify #1 saturation - 50
6. col modify #1 lightness + 20
Room for improvement, needs more desaturation
16.04.2025 06:31 — 👍 53 🔁 9 💬 3 📌 1
AlphaFold as a Prior: Experimental StructureDetermination Conditioned on a Pretrained Neural Network https://www.biorxiv.org/content/10.1101/2025.02.18.638828v1
22.02.2025 02:49 — 👍 8 🔁 5 💬 0 📌 1
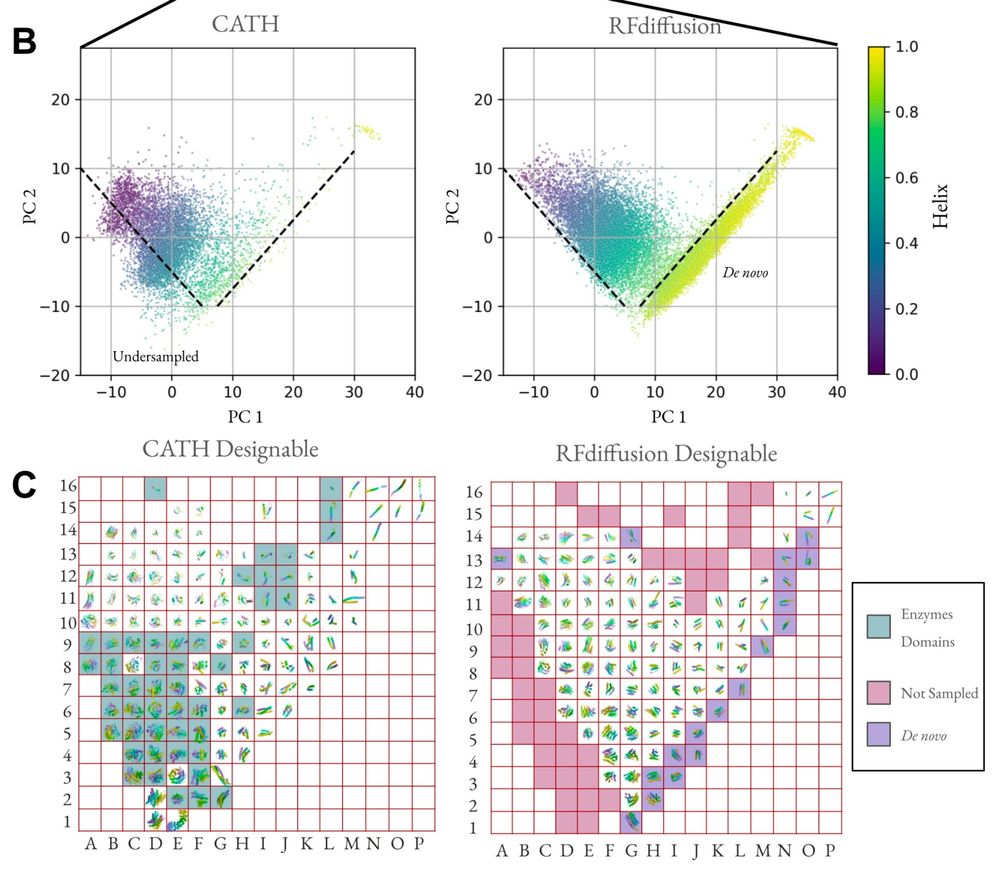
Protein structure embeddings reveal undersampled and de novo structure space. (B) First two principal components of mean-pooled ESM3 embeddings colored by helix content determined by DSSP. The indicated dashed guide lines denote visual boundaries of native structure space not sampled (Undersampled) and novel regions of protein structure for this space only observed in samples but not in native structures (De novo). (C) Rasterized visualization of panel B with 16 equally spaced grid squares in each principal component axis. A representative structure from each grid was chosen at random. Empty grid squares indicate the absence of any structure in the enclosed region. De novo alpha helices are shaded along the lower-right diagonal and the structures from CATH which do not have corresponding structures in the samples are shaded along the left and top rims. The structures are displayed in CATH raster plot are given in the Supplementary Information.
Protein backbone diffusion models consistently undersample catalytically important motifs, and oversample idealized helices, raising questions about the appropriateness of these methods in designing starting points for enzyme design & evolution. From www.biorxiv.org/content/10.1...
19.01.2025 16:40 — 👍 86 🔁 16 💬 2 📌 0

We processed the results of the BindCraft user experience poll and we are quite happy with how it turned out. We had over 60 responses from many different users, turns about about a quarter are from industry and a quarter of users run it via Google Colab!
19.01.2025 12:15 — 👍 17 🔁 5 💬 1 📌 0
Off-axis electron holography of unstained bacteriophages: Toward electrostatic potential measurement of biological samples pubmed.ncbi.nlm.nih.gov/39818354/
18.01.2025 02:59 — 👍 3 🔁 1 💬 0 📌 1

iAPEX: Improved APEX-based proximity labeling for subcellular proteomics using an enzymatic reaction cascade
Ascorbate peroxidase (APEX) is a versatile labeling enzyme used for live-cell proteomics at high spatial and temporal resolution. However, toxicity of its substrate hydrogen peroxide and background la...
Proud to share our work spear-headed by PhD student Tommy Sroka with major contributions by #xenopus expert Kerstin Feistel.
Our iAPEX #ProximityLabeling method for #MassSpec based subcellular #proteomics works by locally generating H2O2 using a D-amino acid oxidase that activates APEX2 in situ.
11.01.2025 19:51 — 👍 34 🔁 16 💬 1 📌 1
In a great collaboration with @hummerlab.bsky.social and the Kräusslich lab: HIV capsid doesn't break at the NPC; instead, it cracks open the NPC itself! Details in Cell: authors.elsevier.com/sd/article/S... @mpibp.bsky.social @uniheidelberg.bsky.social A thread below:
17.01.2025 18:43 — 👍 428 🔁 136 💬 7 📌 22

Opening slide for state of the lab PowerPoint presentation.
Gave our first state of the lab address this week. I broke down the cost of science, from people to stuff. Dove into spending compared to budgets. Spent time on grant submissions and manuscript/project expectations for the year.
I want to be as transparent as possible & this was a great start! 🧪🧠👩🔬
18.01.2025 16:12 — 👍 36 🔁 2 💬 3 📌 1

Scientific Director, Cal-Cryo@QB3, UC Berkeley’s Cryo-EM Facility
University of California, Berkeley is hiring. Apply now!
Scientific Director, Cal-Cryo. We are looking for an outstanding microscopist to run our cryo-EM facility (two Krioses and an Arctica), increase training, engagement, and collaboration on campus, expand our user base, and join our community of cutting edge cryo-EM researchers. tinyurl.com/ykasna3p
15.01.2025 21:53 — 👍 23 🔁 26 💬 0 📌 4
If international people are keen to come to the UK then York would love to host them for a Newton Fellowship:
royalsociety.org/grants/newto...
We have brilliant centres for structural biology, chemical biology, parasitology, haematology, atmospheric science, and much more
16.01.2025 21:37 — 👍 11 🔁 7 💬 1 📌 1

New development in #cryoEM specimen preparation from the Efremov lab that enables structure determination from a single plate of cells. Will be exciting to see how this revolutionizes low copy structure determination and investigations from native sources.
www.biorxiv.org/content/10.1...
15.01.2025 19:33 — 👍 11 🔁 5 💬 0 📌 0
Full disclosure, at risk of revealing the full extent of my ineptitude, the last time I performed motion correction in RELION with a combination of LZW-TIFF/.gain, I merely renamed the gain reference to .tiff. It....didn't seem to mess things up.
13.01.2025 11:30 — 👍 1 🔁 0 💬 0 📌 0
I have been similarly confused by this previously. However, I believe the intention behind the phrasing in this section is to say that: relion_convert_to_tiff will output the reciprocal *if* it is necessary--based on the logic in loadEERGain: github.com/3dem/relion/...
13.01.2025 09:42 — 👍 0 🔁 0 💬 0 📌 0
Investigator scientist @MRC-LMB,
GPCR structures and cryoEM method development
Also 🎸🎹🎻🏃♀️🧗♀️🚵♀️📷
CEO and co-founder of turboTEM Ltd. Tenured Associate Professor (Royal Society URF) at Trinity College Dublin and Ultramicroscopy Group leader. School of Physics Director of EDI. EMS Board member.
PI of lab in the areas of protein lipidation and transition metal transport and homeostasis. Interested in how nature does chemistry. Opinion my own and does not represent anyone else.
Group leader, Norwich Medical School (UEA)...
membrane proteins, mitochondria in health and disease, bioenergetics
https://research-portal.uea.ac.uk/en/persons/paul-crichton
News from the Sternberg lab at Columbia University, Howard Hughes Medical Institute.
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
Postdoc @ Alex MacKerell Lab
Former member of Grossfield Lab
Interested in Membrane Permeability, Phase separation, and Computer Aided Drug Discovery.
Currently working with Permeability predictions and Endocannabinoid signaling players
PhD candidate at University of Toronto
Structural Biology (Cryo-EM enthusiast), Medicinal Chemistry
-Obviously not a complete failure-
Membrane protein structural biologist
❤️ photography,bouldering, surfing, travelling, movies and books.
Opinions are my own.
Group leader at MRC Laboratory of Molecular Biology #transcription #splicing #cryoEM
https://www2.mrc-lmb.cam.ac.uk/groups/zhang/#foogallery-82/i:165
don’t tell me you aren’t
✉️ you can also dm me your fav sci memes to post here
Understanding Life In Molecular Detail.
An interdisciplinary research centre at @universityofleeds.bsky.social with over 400 researchers who are passionate about understanding life in molecular detail.
https://astbury.leeds.ac.uk/
Structural Virology unit directed by Félix Rey @pasteur.fr in Paris | @CNRS #UMR3569 | Managed by Rey Lab Members |
Biochemist studying #Cilia, Cell Signaling and Protein Trafficking via Proximity Labeling and Proteomics.
https://www.mick-lab.com
Dpt. for #MolecularSociology @mpibp.bsky.social
https://www.biophys.mpg.de/molecular-sociology
Assistant Professor @ Princeton MolBio |
From transposons to telomeres and back | bicycles and biochemistry
https://ghanim.lab.princeton.edu
We are a cryoEM-focused US-based CRO (we own 3 microscopes on the West Coast, and 3 microscopes on the East Coast), and we acquired Proteos in Kalamazoo, MI, in January of this year. Contact us for cryoEM, gene-to-structure, or protein production.
PhD student in the Grinter Lab at Monash University + The University of Melbourne. Interested in CryoEM, de novo protein design and host-pathogen interactions. Views my own. 🦠🔬🌈
PhD Student 👨🏻🎓 Microbiologist & Structural biologist 🔬🦠
Fronzes Team - CNRS UMR 5234 - Université de Bordeaux 🇫🇷
fronzeslab.cnrs.fr @univbordeaux.bsky.social
#microsky #cryoEM #cryoET