
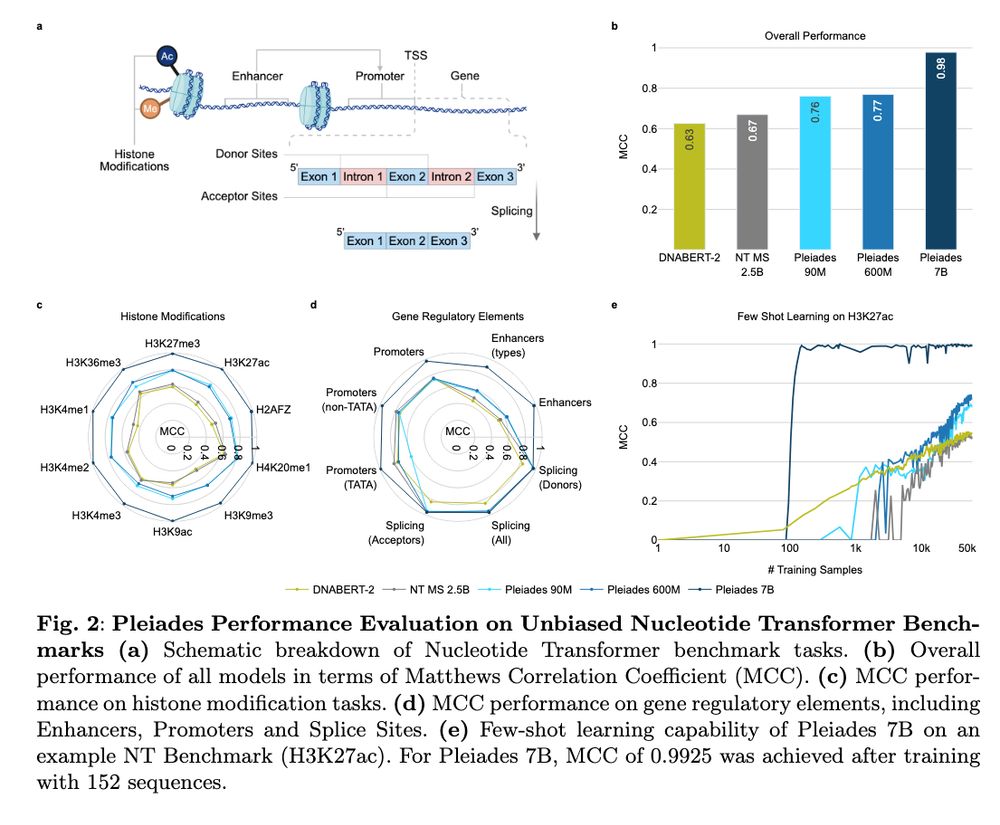
Human whole epigenome modelling for clinical applications with Pleiades www.biorxiv.org/content/10.1... 🧬🖥️🧪
07.08.2025 18:30 — 👍 1 🔁 1 💬 0 📌 0
Thanks everyone involved in the work. Special thanks also to our amazing team of collaborators: @mercedesbarrero.bsky.social, Payer lab and Menendez lab
18.07.2025 18:30 — 👍 1 🔁 0 💬 0 📌 0
Thrilled to share with you the latest work from my PhD in Bigas lab at @researchmar.bsky.social @carrerasijc.bsky.social published in @bloodjournal.bsky.social
We’ve programmed blood stem cells from PSCs using CRISPRa screen and found a new “bloody” cocktail
#CRISPR #HSCs #StemCells #CellFate
18.07.2025 18:28 — 👍 3 🔁 0 💬 0 📌 1

(1/n) Thread @matteomazzocca.bsky.social @domenicnarducci.bsky.social Simon Grosse-Holz @jessematthias.bsky.social preprint
Q: how does chromatin move?
Using MINFLUX, SPT & SRLCI, we track chromatin dynamics across 7 orders of magnitude in time to provide answers www.biorxiv.org/content/10.1...
15.05.2025 12:37 — 👍 69 🔁 22 💬 2 📌 1

Emerging roles for the nucleolus in development and stem cells
Summary: This Review explores roles of the nucleolus in mammalian development, with a particular focus on genome organisation and chromatin regulation.
Nice review about alternative roles of nucleolus in genome organization in stem cells
Emerging roles for the nucleolus in development and stem cells | Development
#Review #Heterochromatin #GenomeOrganization #StemCells
journals.biologists.com/dev/article/...
15.05.2025 12:42 — 👍 0 🔁 0 💬 0 📌 0
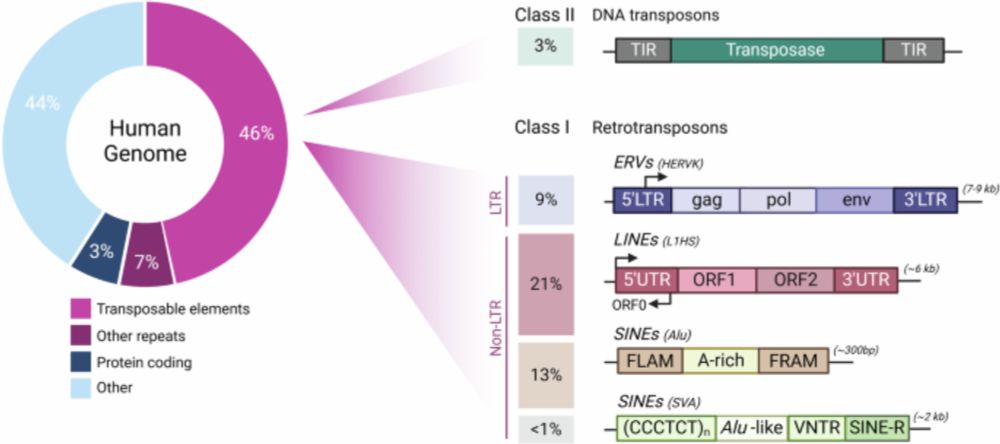
Transposable elements as genome regulators in normal and malignant haematopoiesis - Blood Cancer Journal
Blood Cancer Journal - Transposable elements as genome regulators in normal and malignant haematopoiesis
I am excited to share our lab's first review out with in Blood Cancer Journal. We discuss the impact of transposable elements on genome regulation and variation in both normal
haematopoietic processes and haematological cancers. www.nature.com/articles/s41...
06.05.2025 16:31 — 👍 27 🔁 10 💬 0 📌 0

Stem cells as role models for reprogramming and repair
Stem cells are a promising source for cellular therapies across many diseases and tissues. Their inherent ability to differentiate into other cell types has been the focus of investigation over decade...
Review about principles of stem cell biology and cellular reprogramming approaches and their use for current and future therapeutic purposes
Stem cells as role models for reprogramming and repair | Science
#Review #StemCells #Reprogramming #RegenerativeMedicine
www.science.org/doi/10.1126/...
01.05.2025 22:43 — 👍 1 🔁 0 💬 0 📌 0
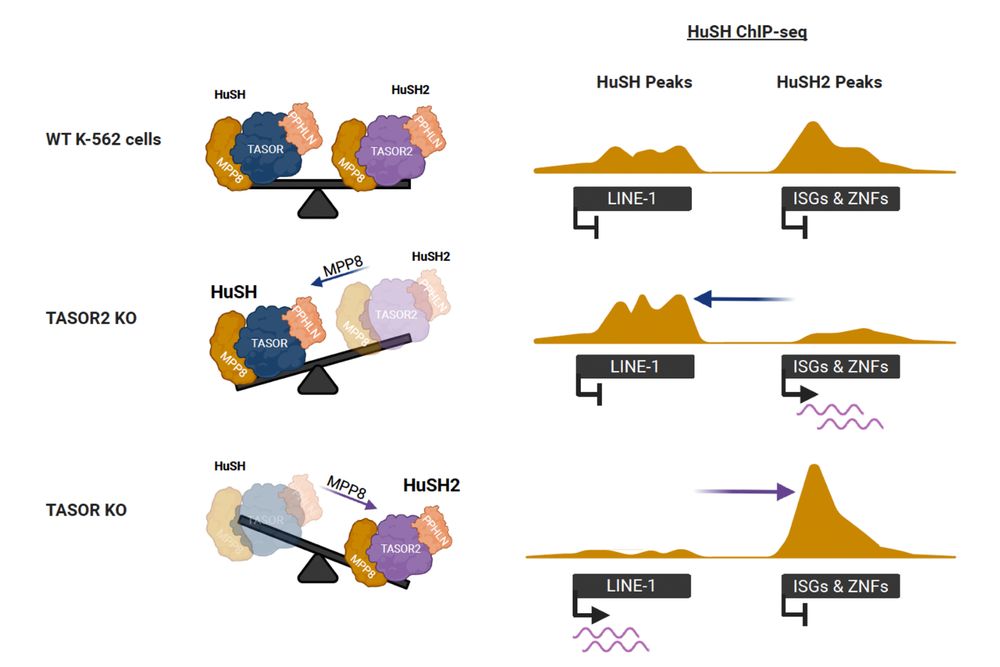
The HuSH complex silences retrotransposable elements. We identify HuSH2, centered on TASOR2, targeting KRAB-ZNFs and interferon genes, balancing retrotransposon silencing and immune regulation. www.nature.com/articles/s41...
03.11.2024 15:20 — 👍 76 🔁 38 💬 5 📌 0

Predictable Engineering of Signal-Dependent Cis-Regulatory Elements
Our latest: uncovering hierarchical design rules for cis-regulatory elements with a high-throughput screen
CRE activity follows multiplicative modular rules, tuned by spacing & positioning
Enables engineering synthetic CREs for predictable tissue patterning
www.biorxiv.org/content/10.1...
10.03.2025 04:19 — 👍 95 🔁 27 💬 3 📌 2
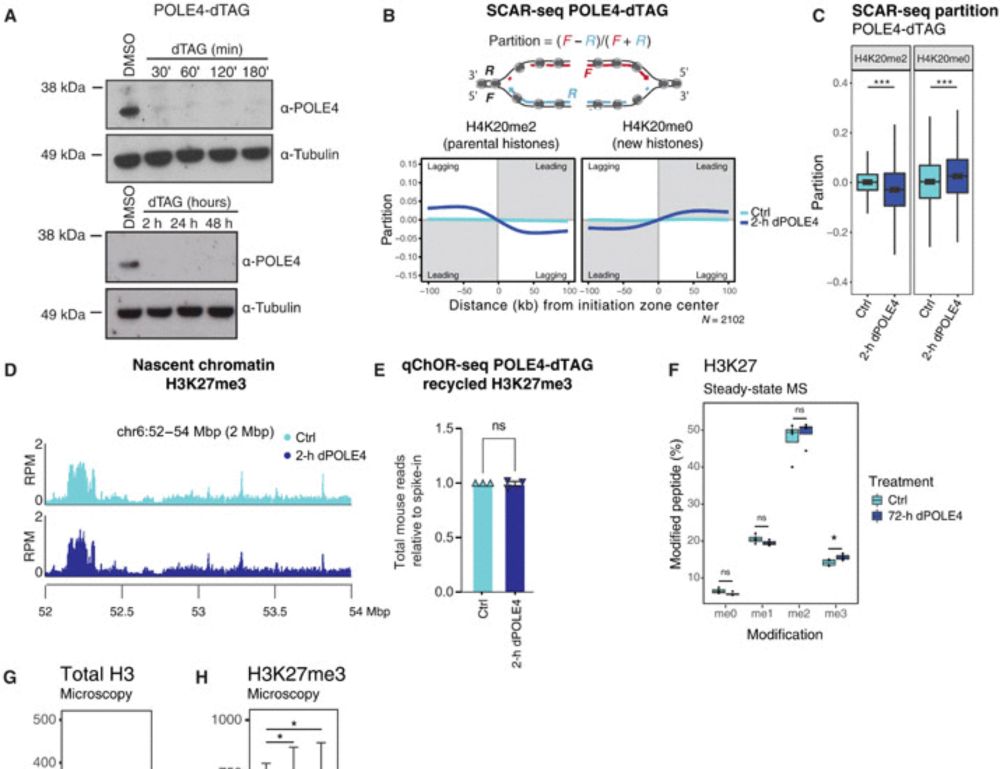
Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability
Losing parental histones during DNA replication fork passage challenges differentiation competence and cell viability.
𝐇𝐨𝐰 𝐝𝐨 𝐜𝐞𝐥𝐥𝐬 𝐫𝐞𝐦𝐞𝐦𝐛𝐞𝐫 𝐰𝐡𝐨 𝐭𝐡𝐞𝐲 𝐚𝐫𝐞 𝐚𝐟𝐭𝐞𝐫 𝐃𝐍𝐀 𝐫𝐞𝐩𝐥𝐢𝐜𝐚𝐭𝐢𝐨𝐧? Our new study “Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability” is out in 𝘚𝘤𝘪𝘦𝘯𝘤𝘦 𝘈𝘥𝘷𝘢𝘯𝘤𝘦𝘴. Led by @lleonie.bsky.social @biranalva.bsky.social 🧵 More below👇
19.02.2025 19:35 — 👍 112 🔁 53 💬 5 📌 5

RNA polymerase II coordinates histone deacetylation at active promoters
Transcription initiation limits histone acetylation and H2AZ incorporation at promoters.
Histone modifications occur independently of transcription
RNA polymerase II coordinates histone deacetylation at active promoters | Science Advances
#chromatin #transcription #generegulation
www.science.org/doi/10.1126/...
13.02.2025 17:49 — 👍 10 🔁 3 💬 0 📌 0
Scientist/PI Hubrecht Institute. Exploring gene regulation in early development and cancer. Single-cell spatial genome organization & epigenomics
Contributing to a better world through research into the fundamental processes of living cells, tissues and organisms. The Hubrecht Institute is a research institute of KNAW and is affiliated with the UMC Utrecht.
Post-Doc @hubrechtinstitute.bsky.social @jopkind.bsky.social. PhD @crg.eu PayerLab. Epigenetics, the X and single-cell technologies.
Epigenome Technologies, pioneering in epigenetic and single-cell analysis, offers advanced assay kits and optimization services.
PhD Student at @belverlab.bsky.social in #Leukemia | MSc Medicinal Chemistry (Toulouse UT3) | 🇦🇩
Bio-protocol (www.bio-protocol.org) was founded in 2011 by a group of researchers at Stanford University with a singular mission: Improving research reproducibility by publishing high-quality protocols in the life sciences.
Gene regulation, enhancer, 3D chromatin, transposable elements, cancer epigenetics, inflammation, leukaemia
ISEH promotes the scientific knowledge & clinical application of basic hematology, immunology, stem cell research, cell and gene therapy.
The American Federation for Aging Research (AFAR) is a national non-profit organization whose mission is to support and advance healthy aging through biomedical research. www.afar.org
Chromatin biochemistry and genomics in development and cancer. UW-Madison School of Medicine and Public Health
TheLewisLab.net
Associate professor at @www.unizar.es
Embryology, Developmental Biology, Epigenetics and Aging Researcher_Vet_2XMum
The mission of the American Association for Cancer Research (AACR) is to prevent and cure cancer through research, education, communication, collaboration, science policy and advocacy, and funding for cancer research.
Delivering the Bay Area’s best journalism every day.
EMBO is the organization of more than 2,100 leading researchers that promotes excellence in life sciences in Europe and beyond.
https://www.embo.org/
Updates from our team at University of Cambridge, on a mission to transform human health through a deep understanding of stem cell biology.
We are the Hospital de Mar Research Institute. An institution dedicated to biomedical research. We are Hospital del Mar, #iCerca and located at the PRBB.
The Anja Groth group in Copenhagen studies chromatin replication, epigenetic memory and (epi)genome stability in the context of mitotic cell division
Dad, husband, teacher, coach, veteran. Governor of Minnesota. Working to move our state forward as #OneMinnesota.





























