
Mascot Newsletter logo
Mascot Newsletter, January 2026:
Ion intensity: peak area vs signal to noise?
Duplexing deuterated water-labeled samples for protein turnover rate estimation.
Role-based access with Mascot Security is useful in a variety of cases.
www.matrixscience.com/nl/202601/ne...
#proteomics #massspec
29.01.2026 09:55 — 👍 0 🔁 0 💬 0 📌 0

Screenshot of Mascot Distiller showing the menu item for cleaning up old cache files.
Proteomics software is prone to filling up the disk with data and temp files, which can cause weird issues. This blog article lists the most common errors in Mascot products and how to free up disk space.
www.matrixscience.com/blog/strange...
#proteomics #massspec
09.01.2026 10:59 — 👍 0 🔁 0 💬 0 📌 0

Mascot Newsletter logo
Mascot Newsletter, December 2025:
NCBI nr FASTA replacement.
Immunoregulation during pregnancy.
Mascot Server as system service on Linux.
www.matrixscience.com/nl/202512/ne...
#proteomics #massspec
18.12.2025 11:50 — 👍 1 🔁 0 💬 0 📌 0

Example MS/MS spectrum of KQLATKAAR using peak area as peak intensity (top) or signal-to-noise as peak intensity (bottom).
When raw data is processed into peak lists, there is a choice: how should peak intensity be calculated? Mascot Distiller defaults to peak area. The alternative, signal-to-noise (S/N), can be useful with detecting diagnostic ions.
#proteomics #massspec
www.matrixscience.com/blog/peak-li...
01.12.2025 16:19 — 👍 1 🔁 0 💬 0 📌 0

Mascot Newsletter logo
Mascot Newsletter, November 2025:
Top-3 quant with DDA-PASEF.
ZooMS peptide markers for South American Xenarthrans.
Migrating from Windows 10 to Linux could extend the lifetime of your hardware.
www.matrixscience.com/nl/202511/ne...
#proteomics #massspec
27.11.2025 09:35 — 👍 1 🔁 0 💬 0 📌 0
Replacements for NCBI nr
The NCBI nr FASTA file of protein sequences is no longer updated. We've summarised instructions for creating it from the BLAST database -- or you may be better off using TrEMBL.
www.matrixscience.com/blog/replace...
#proteomics #massspec
10.11.2025 14:46 — 👍 1 🔁 1 💬 0 📌 0
Mascot search engine | Protein identification software for mass spec data
Mascot software from Matrix Science - identification, characterisation and quantitation of proteins using mass spectrometry data
The free Mascot service at matrixscience.com is currently being hammered by a distributed denial of service (DDoS) attack. The incident started around 11:00 GMT. We have mitigated the attack, which means some Mascot functionality is unavailable for now.
06.11.2025 13:55 — 👍 1 🔁 0 💬 0 📌 0

Mascot Newsletter logo
Mascot Newsletter, October 2025:
Spectrum-centric DIA scoring.
Molecular mechanisms of SBK2 kinase on the heart.
Windows update issue with IIS (resolved).
www.matrixscience.com/nl/202510/ne...
#proteomics #massspec
29.10.2025 15:46 — 👍 1 🔁 0 💬 0 📌 0
Top-3 Quantitation Summary in Mascot Daemon 3
Mascot Daemon can export protein level quant intensities for replicate LFQ, SILAC and iTRAQ/TMT. The latest version can also export intensity values calculated using the "Top-3" method, illustrated here with DDA-PASEF data.
www.matrixscience.com/blog/top-3-q...
#proteomics #massspec
17.10.2025 14:52 — 👍 0 🔁 0 💬 0 📌 0
New Mascot distributor in India: BioInnovations
We are pleased to announce a new distributor for Matrix Science products! Customers in India can now buy Mascot Server through BioInnovations, who provide products and services for life sciences and analytical and clinical labs. #proteomics #massspec
www.matrixscience.com/whats_new/ne...
18.09.2025 09:18 — 👍 0 🔁 0 💬 0 📌 0

Illustration of a DIA MS/MS spectrum with unidentified peaks (top) and labelled peaks for two identified peptides (middle and bottom).
Probabilistic scoring of DIA matches without spectral deconvolution? Not only is it possible, we're working on implementing it in Mascot DIA.
www.matrixscience.com/blog/probabi...
#proteomics #massspec
08.09.2025 09:03 — 👍 5 🔁 4 💬 0 📌 0

Mascot Newsletter logo
Mascot Newsletter, August 2025:
Precursor detection for spectrum-centric DIA.
Reconstructing mediaeval monastic diets.
Microsoft has announced the end of Windows 10 support; your Mascot Server may be affected.
www.matrixscience.com/nl/202508/ne...
#proteomics #massspec
19.08.2025 08:12 — 👍 0 🔁 0 💬 0 📌 0
1. Precursor detection is performed before the database search.
2. The algorithm tries pairing up ions with no prior knowledge of which ones are the complementary pairs. It's a "brute force" approach with a few shortcuts to speed up the computation.
19.08.2025 08:06 — 👍 1 🔁 0 💬 0 📌 0

MS/MS spectrum of RPQYSNPPVQGEVMEGADNQGAGEQGR zoomed in to the region 1150-1750Da. Several complementary b and y ion pairs are labelled.
Peptide identification in spectrum-centric DIA depends on accurate precursor mass. The upcoming version of Mascot Distiller includes a novel algorithm that deduces precursor masses from MS/MS scans.
#proteomics #massspec
www.matrixscience.com/blog/youve-g...
29.07.2025 10:03 — 👍 0 🔁 0 💬 1 📌 0

Mascot Newsletter logo
Mascot Newsletter, July 2025:
Overview of spectrum-centric DIA and Mascot DIA.
Metaproteomics of bacterioplankton communities.
New pricing tiers for Mascot Server, and price reduction when updating from an old version.
www.matrixscience.com/nl/202507/ne...
#proteomics #massspec
25.07.2025 09:24 — 👍 0 🔁 0 💬 0 📌 0
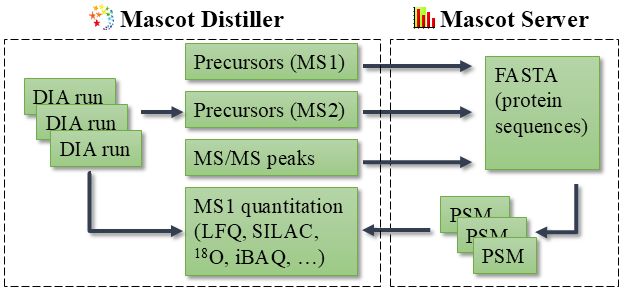
Diagram of Mascot DIA: Mascot Distiller processes DIA runs into peak lists and detects precursor masses in MS1 and MS2 scans; these are sent to Mascot Server for database searching; finally, Distiller downloads the PSMs and performs MS1 quantitation.
We are working on Mascot DIA, which is a universal spectrum-centric solution for DIA analysis. What is spectrum-centric searching meant to solve? Find out in our blog:
www.matrixscience.com/blog/the-pro...
#proteomics #massspec
01.07.2025 15:47 — 👍 2 🔁 2 💬 0 📌 0

Mascot Newsletter
Mascot Newsletter, June 2025:
Annual user meeting summary and preview of Mascot DIA.
ASMS posters using Mascot: forensics, cancer research, food science, fish, ...
Matrix Science posters on LFQ and machine learning.
www.matrixscience.com/nl/202506/ne...
#proteomics #massspec
19.06.2025 08:00 — 👍 0 🔁 0 💬 0 📌 0
Come see our poster ThP 265 today at #ASMS2025: Machine learning-enabled Mascot Server API improves
LFQ in client software like Thermo Proteome Discoverer
05.06.2025 11:43 — 👍 1 🔁 0 💬 0 📌 0

Photo of pens, notebooks and promotional bags.
Pick up a free Matrix Science pen, bag or notebook from #ASMS2025 booth #334 -- while stocks last!
04.06.2025 16:12 — 👍 0 🔁 0 💬 0 📌 0
Come see our poster TP 505 today at #ASMS2025 : Customizable machine learning adapter improves LFQ in Mascot Distiller
03.06.2025 16:18 — 👍 0 🔁 0 💬 0 📌 0

Matrix Science Breakfast Meeting, Monday 2nd June:
Using MS2Rescore to boost identification rates in complex samples,
Tim Van Den Bossche, Computational Omics and Systems Biology Group (CompOmics), Ghent University/VIB-UGent Center for Medical Biotechnology
Mascot DIA: Universal spectrum-centric approach,
Ville Koskinen, Matrix Science
Mascot DIA: Thermo LFQ example,
Patrick Emery, Matrix Science
Register now for the Matrix Science Breakfast Meeting at #ASMS2025 on 2nd June: matrixscience.com/workshop.html
There is no charge for attending the meeting, but advance registration is required. #proteomics #MassSpec
30.05.2025 14:18 — 👍 3 🔁 1 💬 0 📌 0

Count of protein hits and peptide matches, comparing Percolator node (6785 proteins) to Mascot ML (6899 proteins).
When you refine results with Mascot Server and import into Thermo Proteome Discoverer, you should disable MudPIT scoring. This makes the Protein FDR Validator node much happier.
www.matrixscience.com/blog/identif...
#proteomics #MassSpec
29.05.2025 14:53 — 👍 0 🔁 0 💬 0 📌 0

Matrix Science Breakfast Meeting, Monday 2nd June:
Using MS2Rescore to boost identification rates in complex samples,
Tim Van Den Bossche, Computational Omics and Systems Biology Group (CompOmics), Ghent University/VIB-UGent Center for Medical Biotechnology
Mascot DIA: Universal spectrum-centric approach,
Ville Koskinen, Matrix Science
Mascot DIA: Thermo LFQ example,
Patrick Emery, Matrix Science
Register now for the Matrix Science Breakfast Meeting at #ASMS2025 on 2nd June: matrixscience.com/workshop.html
There is no charge for attending the meeting, but advance registration is required. #proteomics #MassSpec
22.05.2025 14:55 — 👍 1 🔁 0 💬 0 📌 0

Mascot Newsletter
Mascot newsletter, May 2025:
ASMS User Meeting registration is open.
Oxygen metabolism in horses (@castiglionelab.bsky.social).
Big databases and large peak lists.
www.matrixscience.com/nl/202505/ne...
#proteomics #massspec
20.05.2025 08:35 — 👍 2 🔁 1 💬 0 📌 0

Matrix Science Breakfast Meeting, Monday 2nd June:
Using MS2Rescore to boost identification rates in complex samples,
Tim Van Den Bossche, Computational Omics and Systems Biology Group (CompOmics), Ghent University/VIB-UGent Center for Medical Biotechnology
Mascot DIA: Universal spectrum-centric approach,
Ville Koskinen, Matrix Science
Mascot DIA: Thermo LFQ example,
Patrick Emery, Matrix Science
Register now for the Matrix Science Breakfast Meeting at #ASMS2025 on 2nd June: matrixscience.com/workshop.html
There is no charge for attending the meeting, but advance registration is required. #proteomics #MassSpec
15.05.2025 15:04 — 👍 3 🔁 0 💬 0 📌 0

Screenshot of the format controls in Protein Family Summary, showing a dropdown item for example_ML_adapter.py
Mascot Server ships with MS2Rescore, which is one of many predictive ML tools for proteomics. Follow this new tutorial if you want to add an integration to a different AI/ML system.
www.matrixscience.com/blog/adding-...
#proteomics #MassSpec
01.05.2025 14:12 — 👍 1 🔁 0 💬 0 📌 0
Mascot help: Agilent instruments
Agilent have published an application note for recommended MassHunter Qualitative Analysis settings when searching DDA peak lists with Mascot Server. We've added it to a new help page for Agilent instruments.
www.matrixscience.com/help/instrum...
#proteomics #MassSpec
24.04.2025 15:23 — 👍 0 🔁 0 💬 0 📌 0

Mascot Newsletter logo
Mascot newsletter, April 2025:
ASMS User Meeting registration is now open.
Competitive displacement of lipoprotein lipase by GPIHBP1.
MS2PIP models for Thermo instruments.
#proteomics #massspec
www.matrixscience.com/nl/202504/ne...
17.04.2025 09:13 — 👍 1 🔁 0 💬 0 📌 0
Big databases and large peak lists
Sometimes you need a very large sequence database, e.g. search all proteobacteria proteins. Mascot Server has no size limit. Search a database of hundreds of millions of protein sequences without running out of resources.
www.matrixscience.com/blog/big-dat...
#proteomics #MassSpec
02.04.2025 14:24 — 👍 0 🔁 1 💬 0 📌 0
Interested in LC-MS -omics (proteomics, metabolomics, lipidomics, exposomics).
Exploring biology with mass spectrometry
Asst Prof of Pharmaceutical Sciences #UCIrvine | Studying #microprotein and #peptide biology in cancer and metabolism | #FirstGen | #LAnative | 🇵🇸🇲🇽-🇺🇸 | https://faculty.sites.uci.edu/martinezlab/
Research associate focusing on data analysis at UNC proteomics core
Research Investigator at U of M. Interested in proteomics, etc. Developer of FragPipe, MSFragger, IonQuant, etc.
Professor at Greater Bay Area Institute of Precision Medicine, Fudan University | Glycan/Protein/RNA/Mass Spectrometry | https://glycoxielab.org/
Proteomics, Mass Spectrometry
Career Scientist in Quantitative Health Sciences at the Mayo Clinic and Proteome Software, into proteomics technologies, mass spectrometry & bioinformatics (he/him)
enthusiastic archaeologist
post doctoral researcher | Collège de France
associated member of TRACES | Université Toulouse Jean Jaurès
Head of Data Science & Engineering at Talus Bio. I post about data science, BioML, AI drug discovery, Python, mass spec, proteomics, and silly things my kids do.
I also blog sometimes: https://willfondrie.com/post
We are using mass spectrometry to study the dynamic proteome in health and disease
Head of Machine Learning at MSAID ✨
Computational proteomics 💻🔬 He/him
4. year, 1. Gen PhD student #UniWürzburg in the Warscheid lab. Phospho-/Proteomics, python, mechanical stress, #TeamMassSpec.
PhD candidate | Proteomics | Bioinformatics | Precision medicine
@kusterlab
Mass spectrometry, proteomics, Midwesterner. Views are my own.
Proteogenomics, computational biology, network biology, regulatory programs in seeds, food allergy, epitopes, bioactive peptides, edible grains. A/Prof at Edith Cowan University.
Doctoral researcher at Bioinformatics Research Group, FH OÖ - Hagenberg | PROTrEIN Marie-Curie early stage researcher | Computational Proteomics | Bioinformatics | Mass spectrometry | she/her
Proteomics data analysis 💻 + levelling up in RimWorld/BG3/coding🤖🙈
Postdoc @Cambridge ➡️ERC Pepinsure
Archaeology, University of Cambridge | The Globe, University of Copenhagen
Ancient Proteins | Medieval Manuscripts | Proteomics and AI | 🇺🇦









