We are thankful to Jay @jshendure.bsky.social for his constructive feedback, and to the funding agency @damonrunyon.org for the support. It's always fun to work with Choi, looking forward to more collaborations in the future.
06.05.2025 19:33 — 👍 2 🔁 0 💬 0 📌 0
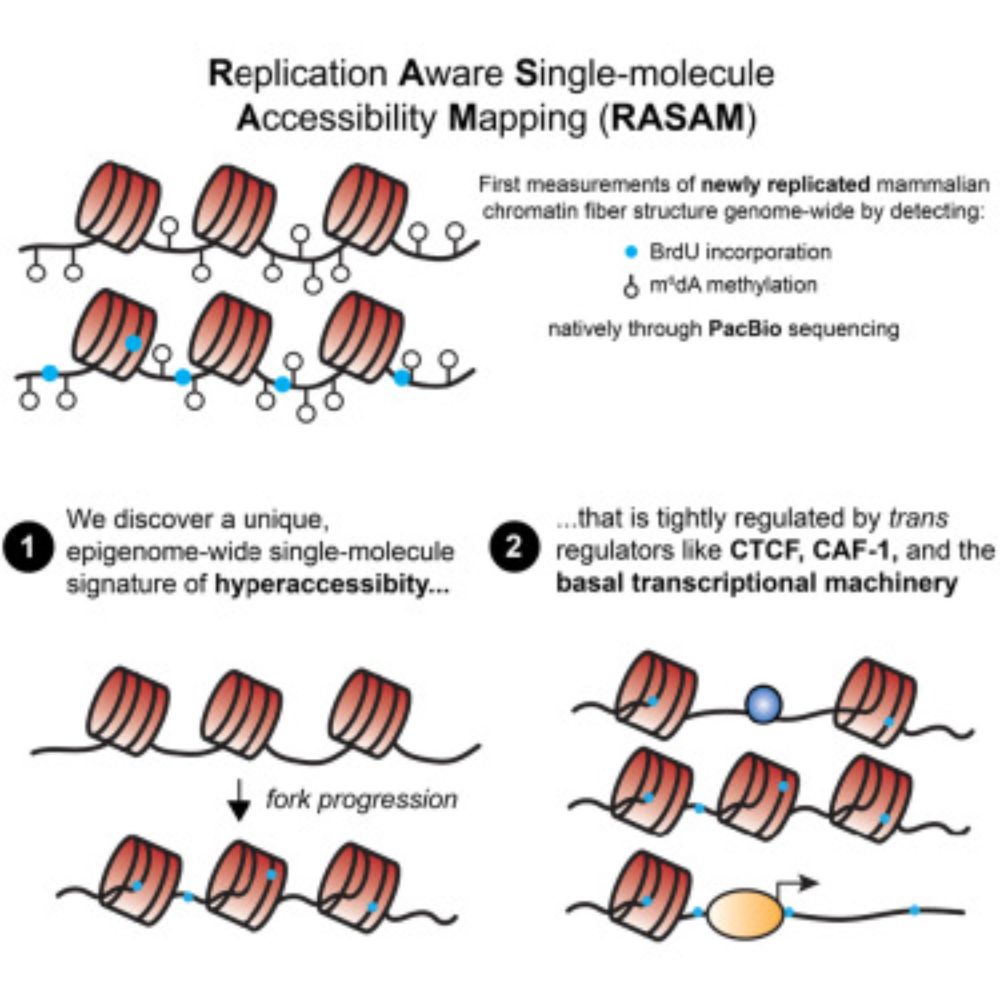
Molecular recording is a exciting and rapidly evolving field. In this perspective, @choijunhong.bsky.social and I delve into the key challenges in the field and highlight a few exciting frontiers that worth more attention.
06.05.2025 19:29 — 👍 5 🔁 1 💬 1 📌 0
Application deadline extended for Taming the BEAST in Beijing!
Interested in #Bayesian #phylogenetics? 🧬
Join the Taming the BEAST workshop — theory + hands-on tutorials incl. #singleCell, #devBio & #lineageTracing!
📍Beijing, July 14–18, 2025
📅 Apply by April 25
I'll teach on TiDeTree & cell trees
🔗 taming-the-beast.org/news/Deadlin...
17.04.2025 19:28 — 👍 9 🔁 5 💬 0 📌 0
Another Monday hype! super excited to see the review + DNA Typewriter/ENGRAM got covered in this nice story.
26.11.2024 01:31 — 👍 1 🔁 0 💬 0 📌 0
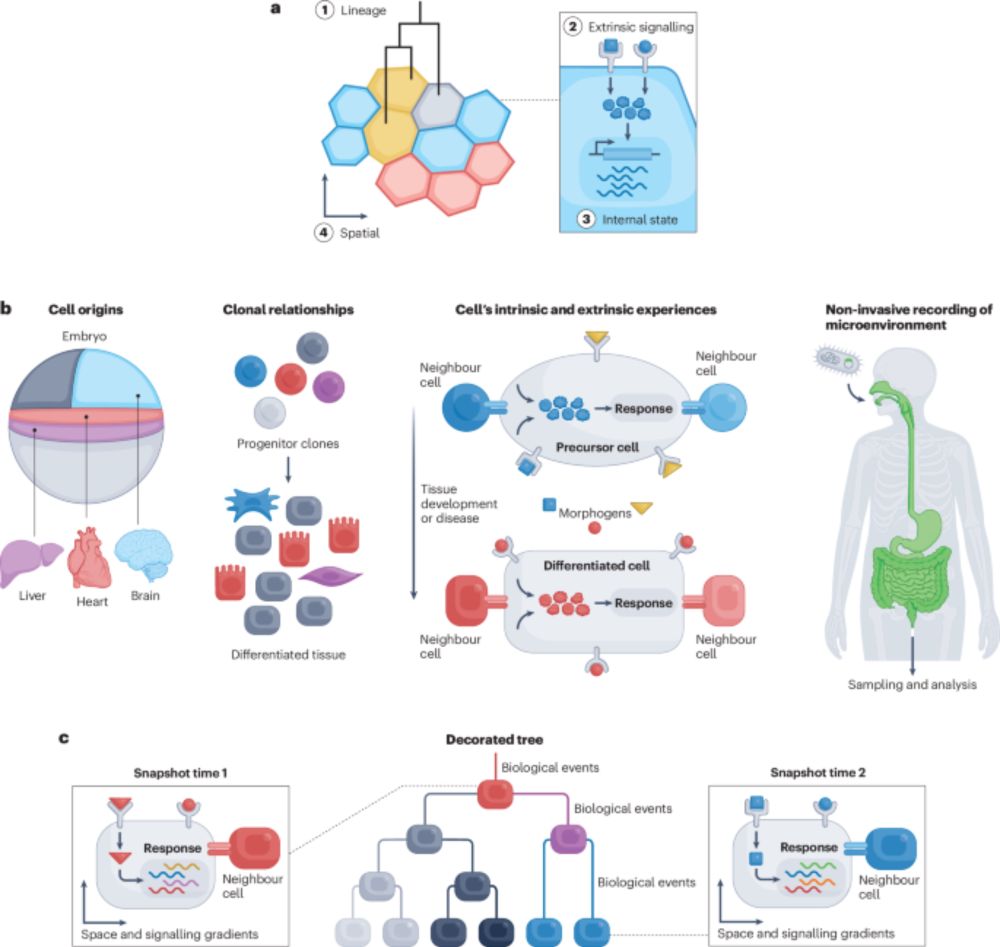
The lives of cells, recorded - Nature Reviews Genetics
Recent advances in genome engineering are enabling the recording of cellular histories into genomes, with single-cell and spatial omics technologies enabling their reconstruction into cellular lineage...
"The lives of cells, recorded"--our new review on genomic recording systems and how they can reveal the dynamics of multicellular development. A pleasure to work on this with amazing colleagues from the Allen Discovery Center for Cell Lineage Tracing.
www.nature.com/articles/s41...
25.11.2024 17:28 — 👍 138 🔁 57 💬 2 📌 4
@DamonRunyon.org Postdoc @UCBerkeleyofficial.bsky.social | Leading Edge Fellow | PhD Princeton | Mobile elements, Structural biology and Genome engineering | http://thawanilab.org
Assistant Professor @UCLA Human Genetics, loves mapping recombination
https://recombination.dev
@DamonRunyon.org Timmerman Traverse Fellow www.sfeirlab.com MSK • PhD in Dewar lab @vanderbilt.edu • from Tbilisi, Georgia 🇬🇪 basic scientist fascinated by mtDNA+🏔️⛷️🎭🗽NYC📍
Postdoc in the Zhang lab @ Broad Institute. Formerly in the Elowitz lab @ Caltech.
Née Ke-Huan Chow.
I ❤️ SynBio and cats.
Banting Postdoc with @JShendure @uwgenome | Prev
@CIHR_IRSC UBC Neuro PhD | Genome engineering, Molecular Recording, neurodevelopment and its disorders | Mountains & skateboarding
Horwitz Lab | Inst for Stem Cell & Regenerative Med | Allen Disc. Center Lineage Tracing | U of Washington | Am I the master of my fate: Am I the Captain of my soul - Asking for a cell
- Posting in a personal manner.
Associate Dean for data science, and Professor of Medicine and Human Genetics at the University of Chicago.
WRF Fellow w/ @jshendure.bsky.social & @leastarita.bsky.social | @uwgenome.bsky.social, @cegs-cmap.bsky.social & @brotmanbaty.bsky.social | PhD w/ @edwardmarcotte.bsky.social | Musician, vinyl collector & Man Utd fan outside the lab. Mostly in that order.
SVP, Head of Analytics & Informatics @ShapeTX | Passionate about gen AI and comp bio research for gene and cell therapy. Formerly @JunoThera & Damon Runyon Fellow w/ @JShendure. CrossFit, Ultimate (frisbee), 🥃 enthusiast, ☕️ aficionado.
Jane Coffin Childs postdoc fellow at Shendure lab at UW
Transcription regulation; deep learning; (bad) developer
Washington Research Foundation fellow in Shendure and Hamazaki labs at UW.
PhD from Robson and Cheng labs at JAX.
Gene editing and Stem cell biology.
Senior editor @science.org. Molecular biology, #DNA, #RNA, #gene regulation, #epigenetics, nuclear biology, #chromatin biology, 3D #genome, #synbio, #CRISPR and gene editing, other bacterial immune systems, and #AI in all these
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Guest Scientist IMP Vienna, Board of Directors NumFOCUS
Incoming Prof UMass Chan Medical
Previously Stanford Genetics, UW CSE.
typing...
(also active on X as @cees_dekker)
Our lab uses experimental and computational methods to design de novo proteins | @Stanford