
@fkma.bsky.social
Thank you Dr. Danny Miller @danrdanny.bsky.social for hosting, and fantastic job Dr. Mastrorosa @fkma.bsky.social!
@uwgenome.bsky.social
brotmanbaty.org/news/long-re...
@fkma.bsky.social
Postdoctoral scholar in the Eichler Lab at UW Genome Sciences. Interested in Mendelian disorders, long-read sequencing and structural variations 🇮🇹 🇪🇺

@fkma.bsky.social
Thank you Dr. Danny Miller @danrdanny.bsky.social for hosting, and fantastic job Dr. Mastrorosa @fkma.bsky.social!
@uwgenome.bsky.social
brotmanbaty.org/news/long-re...
@eichlerlab.bsky.social @glennislogsdon.bsky.social
18.07.2025 17:47 — 👍 0 🔁 0 💬 0 📌 0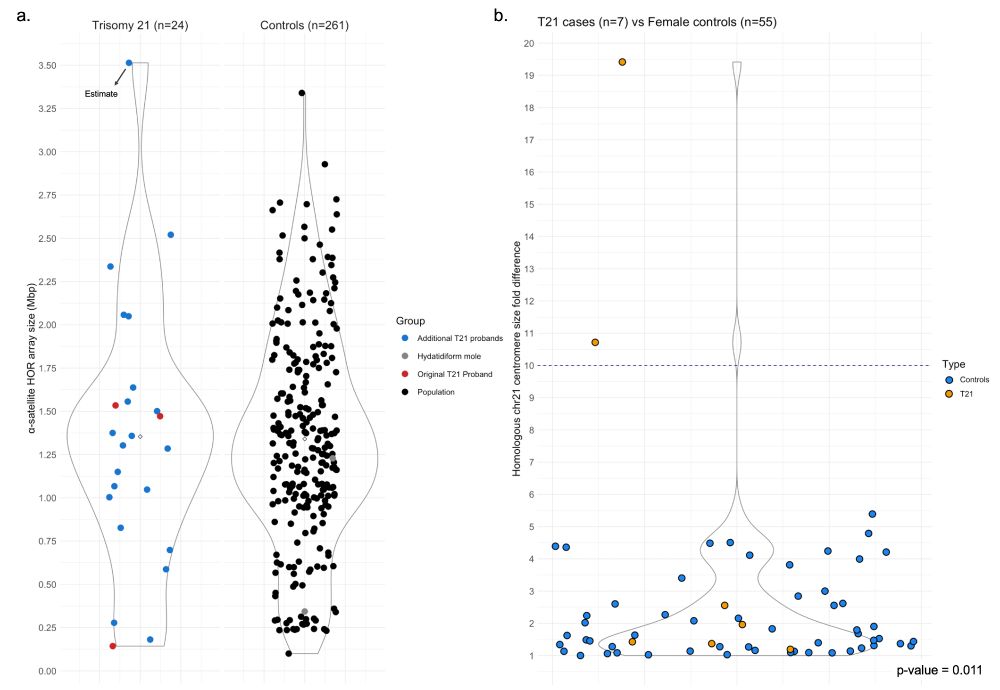
We updated our preprint describing chr21 centromere genetics/epigenetics in families with Down syndrome and general population! We found transgenerational methylation changes in a subset of families and that centromere size asymmetry is exclusive to T21! www.biorxiv.org/content/10.1...
18.07.2025 17:47 — 👍 0 🔁 1 💬 1 📌 0A deep analysis of the 22q11.2 locus in the human population and affected individuals studying the mechanisms and architectures that predispose to complex rearrangements!
09.07.2025 21:27 — 👍 1 🔁 0 💬 0 📌 0I am very happy to have contributed to the publication of the first high-quality, near-complete Middle Eastern genomes. These data will help population studies, disease gene discovery, and increase population representation in genomic datasets!
27.06.2025 17:50 — 👍 0 🔁 0 💬 0 📌 0The most accurate way so far to study transmission, de novo variants, and recombination!
28.04.2025 18:29 — 👍 0 🔁 0 💬 0 📌 0
Nature research paper: Complete sequencing of ape genomes
https://go.nature.com/3EfWwqk

Here is the tool's GitHub page: github.com/EichlerLab/C...
05.02.2025 17:34 — 👍 0 🔁 0 💬 0 📌 0Great work from @koisland.bsky.social from @glennislogsdon.bsky.social lab, who keeps the tool updated!
05.02.2025 17:22 — 👍 2 🔁 0 💬 1 📌 0
We recently published CDR-Finder, a tool to study hypomethylated regions in centromeres (academic.oup.com/bioinformati...). The latest update allows to use the tool to study any region of the genome! This will be useful for neocentromeres, promoters and any methylation variable locus!
05.02.2025 17:22 — 👍 3 🔁 0 💬 1 📌 0