Initial attempt at replicating in relion (parameters in next post). This is for Aca2-RNA, using a 100k subset of the 2D-classified particles (no prior 3D cleanup).
A 1-class ab initio in relion, then local refinement in relion (1.8deg searches+blush) gives a nominally 3.3Å map; 3.5 Å w/out blush.
26.09.2025 02:28 — 👍 57 🔁 24 💬 1 📌 2
Just tried your HR-HAIR + local refinement pipeline (with tweaked settings based on the pre-print) on a dataset of a small protein complex which did not result in any decent ab-initio reconstructions. HR-HAIR took 19 hours, but damn, result is insane! Many thanks!
17.09.2025 07:39 — 👍 19 🔁 3 💬 1 📌 1
We finally wrote up the paper for LocScale-2.0.
edu.nl/pvhxv
Please try it out. LocScale is evolving & we are actively building in new features and improvements. Your feedback will help us shape the next steps.
Code: cryotud.github.io/locscale/
Work by @alok-bharadwaj.bsky.social and Reinier.
15.09.2025 07:39 — 👍 33 🔁 15 💬 0 📌 0
This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
13.09.2025 00:04 — 👍 258 🔁 81 💬 18 📌 12
This is fantastic!! Very nice work!
15.09.2025 07:19 — 👍 1 🔁 0 💬 0 📌 0
And additionally, many thanks to Wouter Van Putte @pxn001.bsky.social (PUXANO) for providing their fantastic graphene-coated EM grids!!!
02.09.2025 11:49 — 👍 1 🔁 0 💬 1 📌 0
Congrats to everyone involved at Sophie Lucas Lab (especially Blueskyless co-first authors Fanny Lambert and Séverine Wautier), @argenx.com and @savvideslab.bsky.social at @viblifesciences.bsky.social!
02.09.2025 11:41 — 👍 0 🔁 0 💬 1 📌 0

Now published @cp-cellreports.bsky.social! Antibody-mediated TGF-β1 activation for the treatment of diseases caused by deleterious T cell activity: www.cell.com/cell-reports...
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.
02.09.2025 11:25 — 👍 8 🔁 4 💬 2 📌 0
We are excited to announce the first in-cell structure of a LINC complex within a native nuclear envelope at subnanometer resolution!! #In-cell cryo-ET + #atomistic MD simulations! @tomdendooven.bsky.social et al!!
www.biorxiv.org/content/10.1...
05.08.2025 12:41 — 👍 41 🔁 14 💬 2 📌 1
Thanks Piotr!
10.07.2025 08:53 — 👍 1 🔁 0 💬 0 📌 0

Finally out @natcomms.nature.com
"Single-domain antibodies directed against hemagglutinin and neuraminidase protect against influenza B viruses".
Great collab between @savvideslab.bsky.social (we performed #CryoEM) and Xavier Saelens Lab, spearheaded by Arne Matthys.
www.nature.com/articles/s41...
10.07.2025 08:45 — 👍 10 🔁 6 💬 1 📌 0
If this is true, it’s abysmally shortsighted for #NIH. The new tools nurtured by the long series of #CASP challenges probably contribute far more in one year to the scientific and industrial economies than the entire funds that have supported it.
03.07.2025 08:08 — 👍 10 🔁 6 💬 1 📌 0
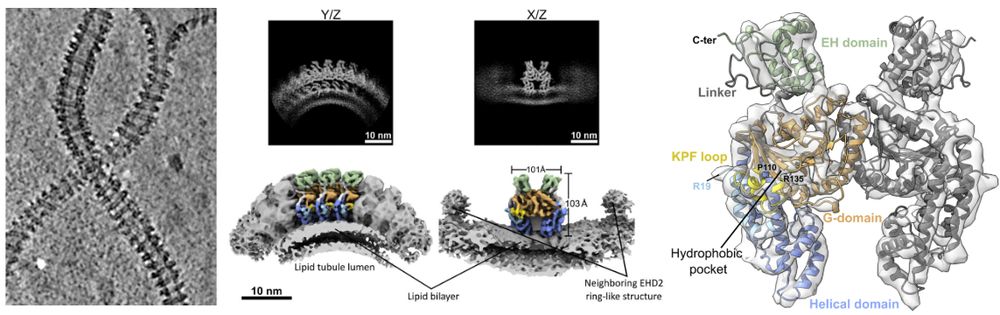
We have a preprint for you
EHD2 forms rings on caveolae necks, in contrast to most EHDs forming helices. We determined its structure on membranes and show that N-term acts as a spacer
By Elena, Vasya @vasiliimikirtumov.bsky.social, Jeff &Oli Daumke www.biorxiv.org/content/10.1...
08.06.2025 11:21 — 👍 107 🔁 34 💬 4 📌 0
We are happy to release LocScale2.0: a tool for context-aware, confidence-weighted cryoEM map optimisation.
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
22.05.2025 10:10 — 👍 121 🔁 38 💬 1 📌 2
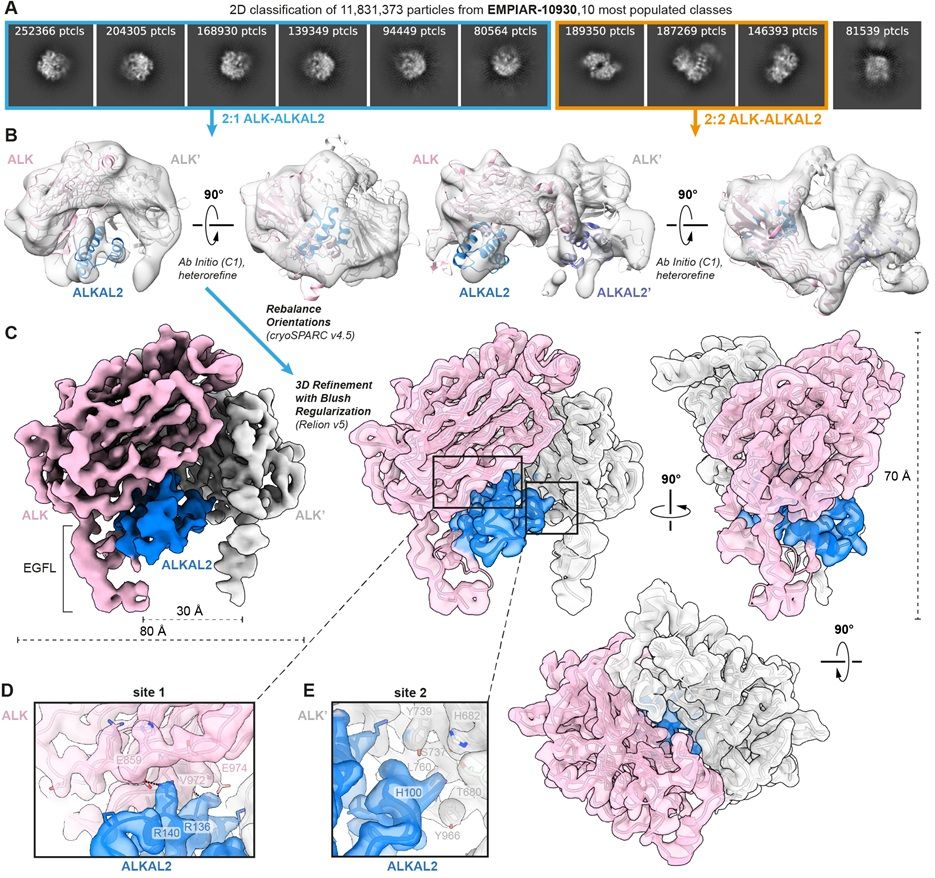
Reanalysis of cryo-EM data from EMPIAR entry EMPIAR-10,930 deposited by Reshetnyak and colleagues, Nature (2021). (A) Ten most populated 2D class averages after performing 2D classification on 11.8 million selected particles. Classes corresponding to 2:1 ALK–ALKAL2 particles are highlighted in blue, while classes corresponding to 2:2 ALK–ALKAL2 particles are highlighted in orange. (B) Ab initio model generation in cryoSPARC results in classes corresponding to both a 2:1 ALK–ALKAL2 and 2:2 ALK–ALKAL2 assembly. Hetero-refinement of the 2:1 and 2:2 ALK–ALKAL2 classes results in the two models displaying 2:1 and 2:2 ALK–ALKAL2 assemblies, respectively. The ALK–ALKAL2 complexes with 2:1 (PDB 7nwz) and 2:2 (PDB 7n00) stoichiometries are shown fitted inside the maps, with ALK colored pink/gray and ALKAL2 colored blue/purple. (C) 3D refinement; a front view is shown of a map sharpened in RELION using a B-factor of −100 Å2, with ALK colored pink/gray and ALKAL2 colored blue. Front, side, and top views are shown of a transparent map, sharpened using deepEMhancer, with fitted structural model.
Reports of diverse stoichiometries have complicated our understanding of the complex between ALK kinase and its #cytokine ligand ALKAL2. @janfelix.bsky.social @savvideslab.bsky.social &co reanalyze #cryoEM data to clarify the situation @plosbiology.org 🧪 plos.io/4lvRfvu
14.04.2025 17:34 — 👍 17 🔁 5 💬 0 📌 0
#CryoSPARC v4.7 is out! 🚀
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
09.04.2025 14:14 — 👍 52 🔁 19 💬 1 📌 2
I'm super happy that our story is now published!
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
21.03.2025 14:09 — 👍 213 🔁 66 💬 7 📌 9
Foam film vitrification for cryo-EM https://www.biorxiv.org/content/10.1101/2025.03.10.642366v1
12.03.2025 15:50 — 👍 19 🔁 9 💬 0 📌 3
Structural biology is in an era of dynamics & assemblies but turning raw experimental data into atomic models at scale remains challenging. @minhuanli.bsky.social and I present ROCKET🚀: an AlphaFold augmentation that integrates crystallographic and cryoEM/ET data with room for more! 1/14.
24.02.2025 12:22 — 👍 137 🔁 60 💬 6 📌 5
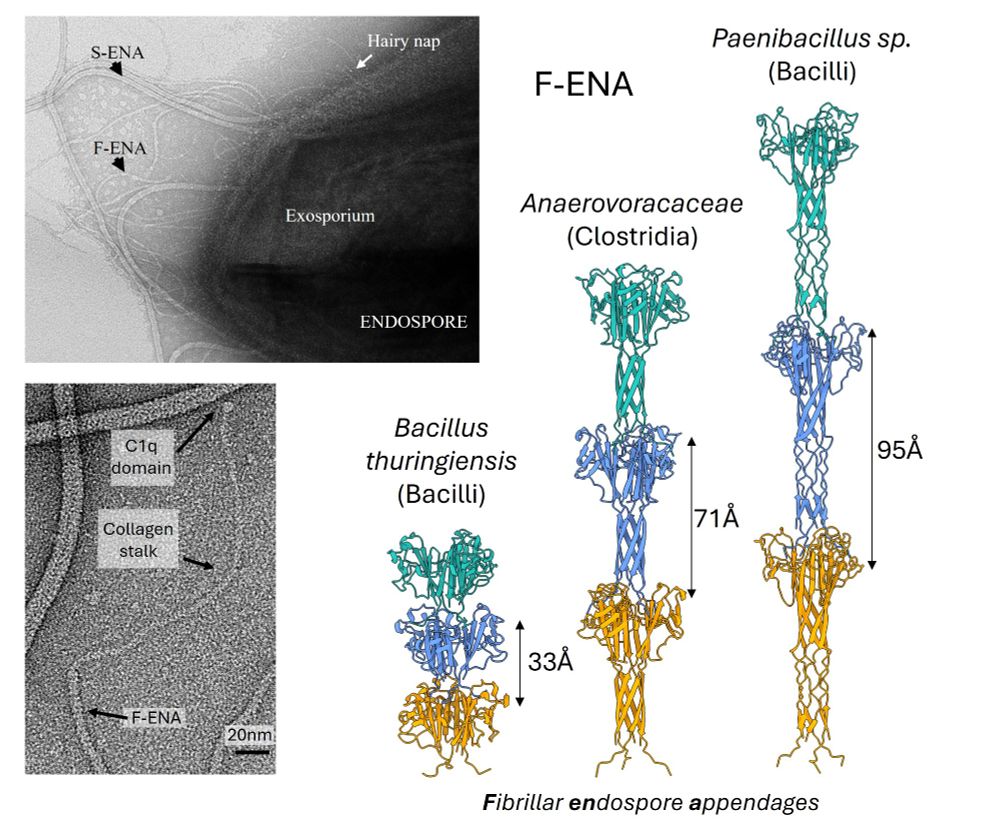
New preprint from the @remaut-lab.bsky.social ! Happy to share our latest work on a new member of the ENA family, protein fibers found on the spore’s surface. Using CryoID @mikesleutel.bsky.social identified F-ENA in BTK. The paper dives deep into its structure analysis by integrating X-ray and AF3
12.02.2025 12:54 — 👍 15 🔁 7 💬 2 📌 2
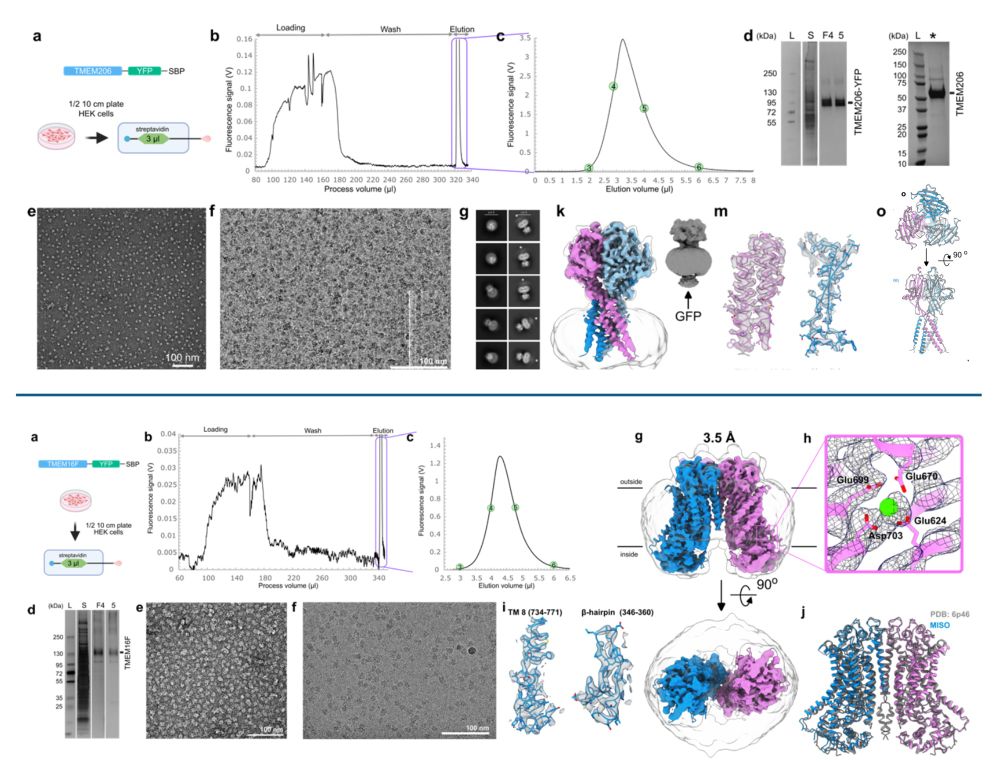
Cryo-EM structures of mammalian membrane proteins from a single cell culture dish?
Yes, Micro ISOlation (MISO) sample preparation makes it possible.
www.biorxiv.org/content/10.1...
Happy to be part of this exciting story and wonderful collaboration with the Rouslan Efremov lab.
16.01.2025 09:27 — 👍 25 🔁 9 💬 1 📌 0
It is definitely a pseudo symmetry case (symmetric dimeric receptor assembly with asymmetric ligand in between). I did not try your suggested workflow. For this particular case 3D orientation rebalancing + BLUSH combo did the trick and results in a nice 3ish Å map from severely anisotropic data.
10.01.2025 16:04 — 👍 1 🔁 0 💬 0 📌 0
To correct my previous post: the map is already deposited to the EMDB, I meant to put the .star file corresponding to the new map on EMPIAR.
10.01.2025 15:16 — 👍 1 🔁 0 💬 0 📌 0
That would be nice! The case is described in our BioRxiv preprint (www.biorxiv.org/content/10.1...) which re-analyses EMPIAR dataset 10930. I made the .star file after 3D refinement with BLUSH available (see suppl. info on BioRxiv). I'm planning to deposit the new map and .star file to EMPIAR later.
10.01.2025 15:07 — 👍 2 🔁 0 💬 2 📌 0
It is nonetheless incredible to see you got a beautiful map from EMPIAR-11918 without BLUSH!
10.01.2025 10:21 — 👍 2 🔁 0 💬 0 📌 0
I just tried your Ab Initio settings followed by NU-refinement for a case where anisotropy was the result of severe preferred particle orientations. While the Ab Initio maps (starting from 3D orientation rebalanced particles) are slightly better, BLUSH gives isotropic maps while NU-ref. does not
10.01.2025 10:19 — 👍 1 🔁 0 💬 2 📌 0
Very interesting! However, the anisotropy of the initial model with standard Ab initio settings seems to be the result of the size of the complex rather than the particle orientation distribution? In cases were a small complex is combined with preferred orientation I think BLUSH > NU-refinement?
10.01.2025 09:11 — 👍 2 🔁 0 💬 2 📌 0
Thanks a lot for the effort! Structural Biologist/CryoEM enthusiast from Gent, BE here. Can I be added as well?
14.11.2024 15:30 — 👍 0 🔁 0 💬 1 📌 0
Structural Biology to the rescue. Postdoc with Gabriel Lander at Scripps Research. PhD with Arne Moeller. #cryoEM ❄️🔬 🎹⛳
Bioinformatics, protein modeling, cryoEM, drug screening, function prediction. Daisuke Kihara, professor of Biol/CS, Purdue U. https://kiharalab.org/ YouTube: http://alturl.com/gxvah
How do bacteria organize their innards?
Associate Prof at UMich
PhD student in the Jakobi lab @Bionanoscience, Delft | Imaging cool things with electrons | cryo-ET
mRNA regulation & decay | biochemistry | structural biology | molecular biology | cancer
All views are personal.
Google Scholar: tinyurl.com/EuValScholar
ORCID: 0000-0002-3721-1739
I like large rocks and small proteins.
BIF fellow doing cryoEM+Protein Design in the Llorca lab ❄️🔬
Structural biologist in the field of innate immunity
https://www.bio.nat.tum.de/en/cryoem/home/
Interested in Structural Biology, deep learning, NextFlow, HPC, viruses, missense mutations archaea, bacteria, evolution & random quirks of nature. (Protein Cosmos feed 🧶🧬) Leads a research group & Structural biology Facility,UNSW Sydney. Opinions my own.
Biochemist/structural biologist who enjoys a bit of maths and programming
Postdoc with Sascha Martens and Tim Clausen at the Vienna BioCenter (VBC). #MSCA Fellow. Formerly University of Oxford and University of Antwerp.
Postdoc at Imperial College
Bacterial conjugation & Cryo-EM
Postdoc at the Francis Crick Institute, previously Cam Biochem PhD.
Structural biology & DNA repair.
When not freezing grids I'm baking, climbing, hiking or taking photos.
Pharmacist | Scientist | Optmisit | Coagulation | Research | I wrote this
Associate Professor @ UW-Madison Biochemistry. www.limlab.org
Telomeres, Cryo-EM, Single-molecule Biophysics. Opinions are my own. 🇸🇬
Assistant Professor, PharmTox, University of Toronto. Signalling, structural biology, protein engineering. 🇨🇦
https://caveneylab.ca
Staff scientist at the university of Osnabrück, exploring membrane transporters by #cryoEM
Postdoc in Delemotte group of Scilifelab and KTH. MD of proteins lipids and sugars.
Getting to grips with chromosome organisation and dynamics using ‘scopes, sequencing and silicon.
Located at @imbavienna.bsky.social - @viennabiocenter.bsky.social
Improving accessibility of TEM and cryo-TEM in research and industry.












