
The new Vienna BioCenter Interdisciplinary Postdoctoral Program (VIP³) is here! We have 8 fully-funded positions for scientists with a PhD in biology, chemistry, physics, bioinformatics etc.
The call is open!
More information: training.vbc.ac.at/post-docs/vi...
#VBCPostDoc
02.03.2026 13:18 —
👍 13
🔁 11
💬 0
📌 1
The Vienna BioCenter PhD program in Molecular BioSciences will offer 25+ fully-funded positions in the Spring Call which is now open. Have a look at our campus: www.vbcphd.at
Institute of Molecular Biotechnology, Max Perutz Labs Vienna, IMP, Gregor Mendel Institute of Molecular Plant Biology
01.03.2026 10:00 —
👍 5
🔁 5
💬 0
📌 1

PhD student recruitment
PhD recruitment information.
Motivated graduates with backgrounds in biological or biomedical sciences, physics, chemistry, mathematics, engineering and/or computer science are invited to apply to our 4-year fully funded PhD programme.
Apply by 17 March 2026.
www.crick.ac.uk/careers-stud...
18.02.2026 10:35 —
👍 18
🔁 10
💬 0
📌 2
We‘re looking for a motivated Master student to join our team!
Do you want to optogenetically control metabolic activity to see how metabolism affects patterning and morphogenesis? 💡🧫🧬🔬
Then please apply!
#optogenetics #metabolism #devbio #hESCs
Please RT. Thank you!🙏
17.02.2026 18:21 —
👍 38
🔁 39
💬 0
📌 0
Best possible start into the fintastic world of zebrafish! 🐠
13.02.2026 19:24 —
👍 2
🔁 0
💬 0
📌 0
We are also recruiting a Postdoc in cell/molecular biology!
3yr, fully funded by @cancerresearchuk.org more details here: news.cancerresearchuk.org/2025/11/27/s...
Apply here: www.imperial.ac.uk/jobs/search-...
23.01.2026 16:23 —
👍 4
🔁 7
💬 0
📌 0

If you want to become an expert in #zebrafish research, apply for the 2026 Zebrafish Development & Genetics course! You will be working in small groups taught by leaders in the field, which can be a transformative experience!
Application deadline: March 2
www.mbl.edu/education/ad...
03.01.2026 09:01 —
👍 8
🔁 7
💬 0
📌 1
🧬 PomBase needs you! 🧬
If you PomBase on a regular basis, now it’s the time to show your support to the team behind this incredibly useful resource (and help them secure funding to keep up the amazing work). Fill out their short user survey, which they will use to support their grant application.
12.11.2025 20:55 —
👍 3
🔁 2
💬 0
📌 0

🌍Open call: Junior Group Leader positions!
Join a world-class biomedical research institute at the heart of the Vienna BioCenter, where curiosity drives discovery.
Lead your own lab, pursue bold ideas, and shape the future of science at the IMP: www.imp.ac.at/career/open-...
10.11.2025 13:26 —
👍 95
🔁 84
💬 1
📌 7

PhD students
Our PhD programme attracts the brightest scientific minds and is an opportunity for talented people to embark on their career in biomedical research.
Motivated graduates with backgrounds in biological or biomedical sciences, physics, chemistry, mathematics, engineering and/or computer science are invited to apply to our 4-year fully funded PhD programme.
Apply by 05 November 2025
www.crick.ac.uk/careers-and-...
02.10.2025 12:53 —
👍 13
🔁 17
💬 0
📌 7
Laura Lorenzo Orts
IMB Mainz
I am excited to announce that I will be moving to IMB Mainz next year! The Winter call for the IPP PhD program is now open; if you are interested in maternal #mRNA regulation and #translation in early vertebrate development, please apply! Deadline: 16 October.
More info: www.imb.de/students-pos...
15.09.2025 12:13 —
👍 66
🔁 25
💬 3
📌 3
Thrilled to announce that I am looking for a Research Technician/Assistant to join my lab @imbavienna.bsky.social. Please consider applying if you’re enthusiastic about #devbio #stemcells #teamwork and excited to help shaping a new lab!
Thankful for any retweet! 🫶
imba.onlyfy.jobs/job/l6u2m980
20.03.2025 12:13 —
👍 50
🔁 42
💬 1
📌 3
Skoglund Lab | Ancient human statistical population genomics
We have a 4-year PhD-student opening in our lab, deadline on Wednesday noon.
For anyone interested in computational genomics of ancient human, pathogen, or canid genomes. www.crick.ac.uk/careers-stud...
If you are curious, send in an application!
17.03.2025 11:01 —
👍 24
🔁 35
💬 0
📌 2

openRxiv has arrived!
We’re thrilled to announce the launch of openRxiv as an independent, researcher-led nonprofit to oversee bioRxiv and medRxiv, the world’s leading preprint servers for life and health sciences.
openrxiv.org/introducing-...
#openRxiv #OpenScience #Preprints #bioRxiv #medRxiv
11.03.2025 13:18 —
👍 407
🔁 231
💬 3
📌 18
Just under two weeks left to apply for PhD positions at the Crick 🔬🧬 Check out this video featuring @sarahwillich.bsky.social for some application tips youtu.be/XjW4LqwKNFs
07.03.2025 11:54 —
👍 3
🔁 4
💬 0
📌 0
Big thanks to Tania for her phospho-expertise and the entire Nurse lab (past and present) for their support, comments and discussions! 🌟
24.02.2025 17:09 —
👍 2
🔁 0
💬 0
📌 0
Without PP2A-B55 or CDC14 cells enter mitosis earlier
This advance is independent of CDK activity regulation. This indicates that in the absence of these Ppases, critical CDK substrates can be phosphorylated at lower CDK activity levels (ie earlier), thus advancing mitotic entry.
24.02.2025 17:09 —
👍 4
🔁 0
💬 1
📌 0
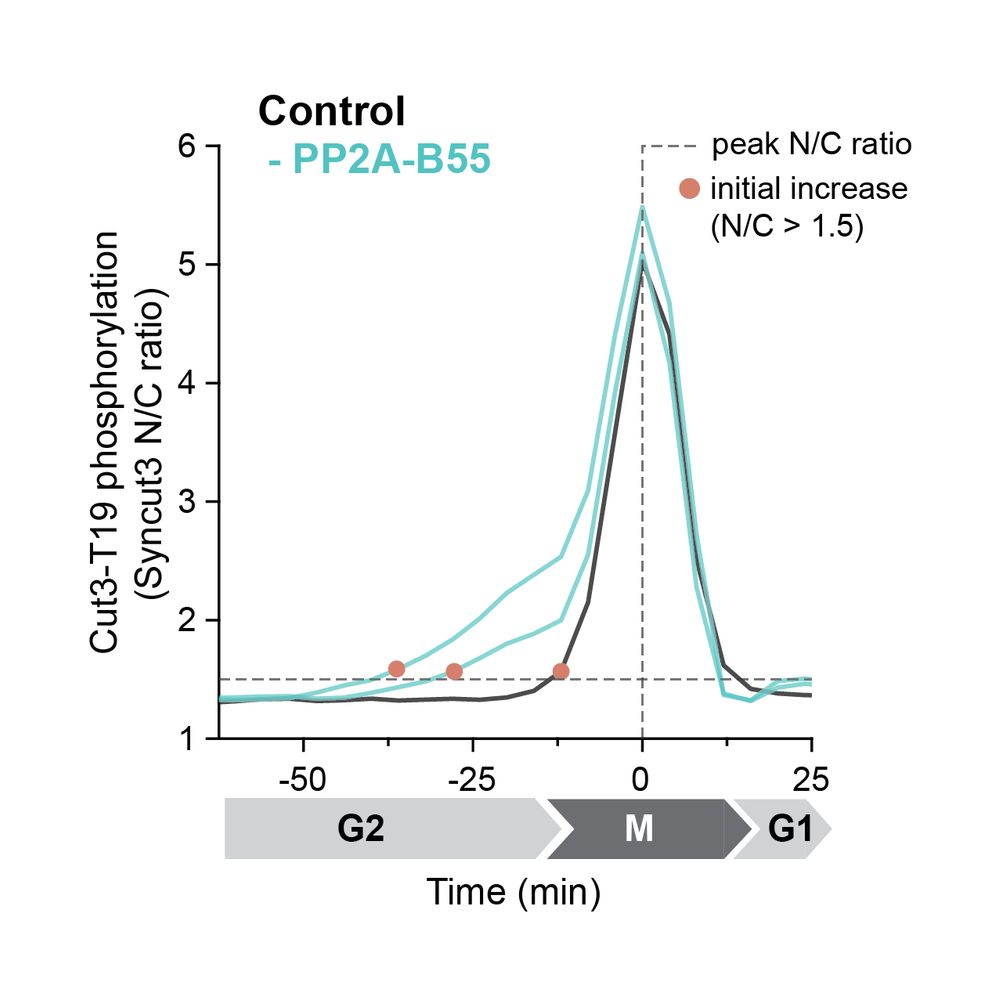
What happens in the absence of the Ppases?
Using a fluorescent sensor, we showed that without PP2A-B55, a candidate substrate (Cut3-T19) is phosphorylated earlier and less rapidly.
24.02.2025 17:09 —
👍 1
🔁 0
💬 1
📌 0
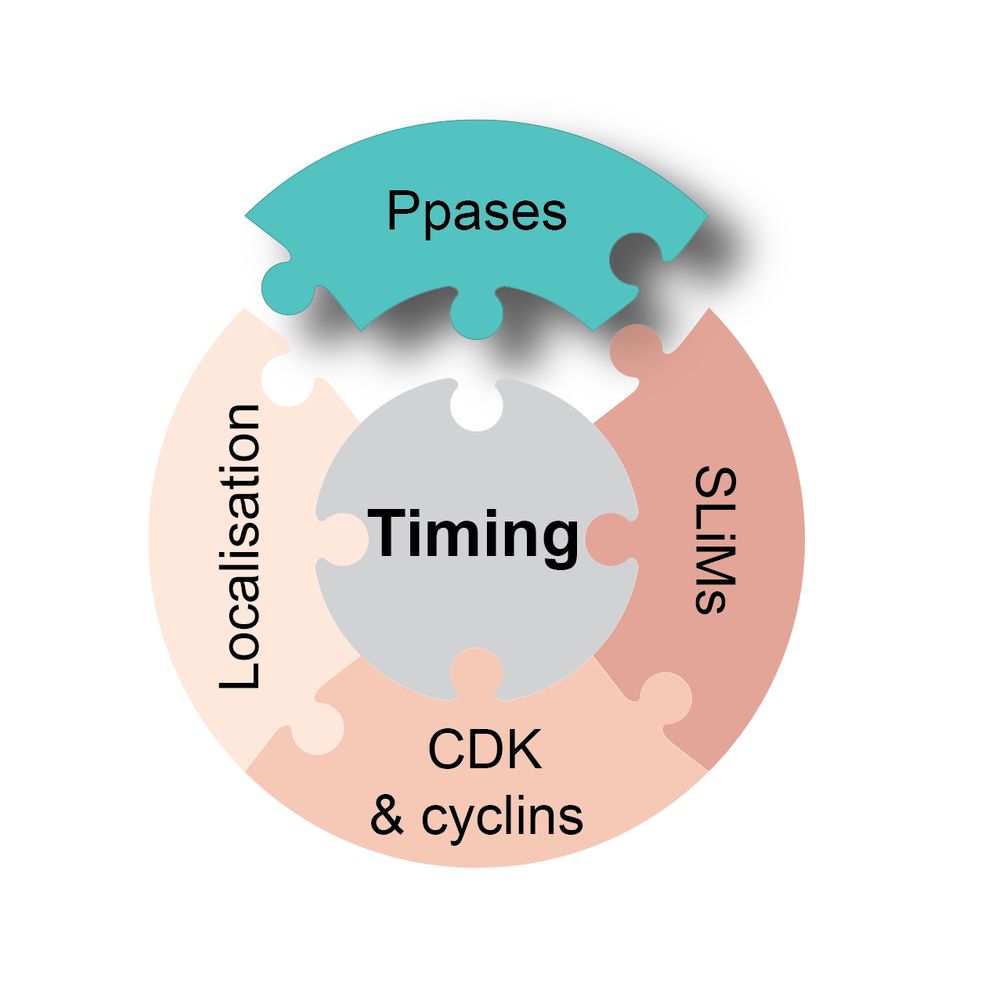
This suggests that the identity of the Ppase that targets a CDK substrate site affects its phosphorylation timing. The exact phosphorylation timing of a given site is determined by a combination of factors.
24.02.2025 17:09 —
👍 1
🔁 0
💬 1
📌 0
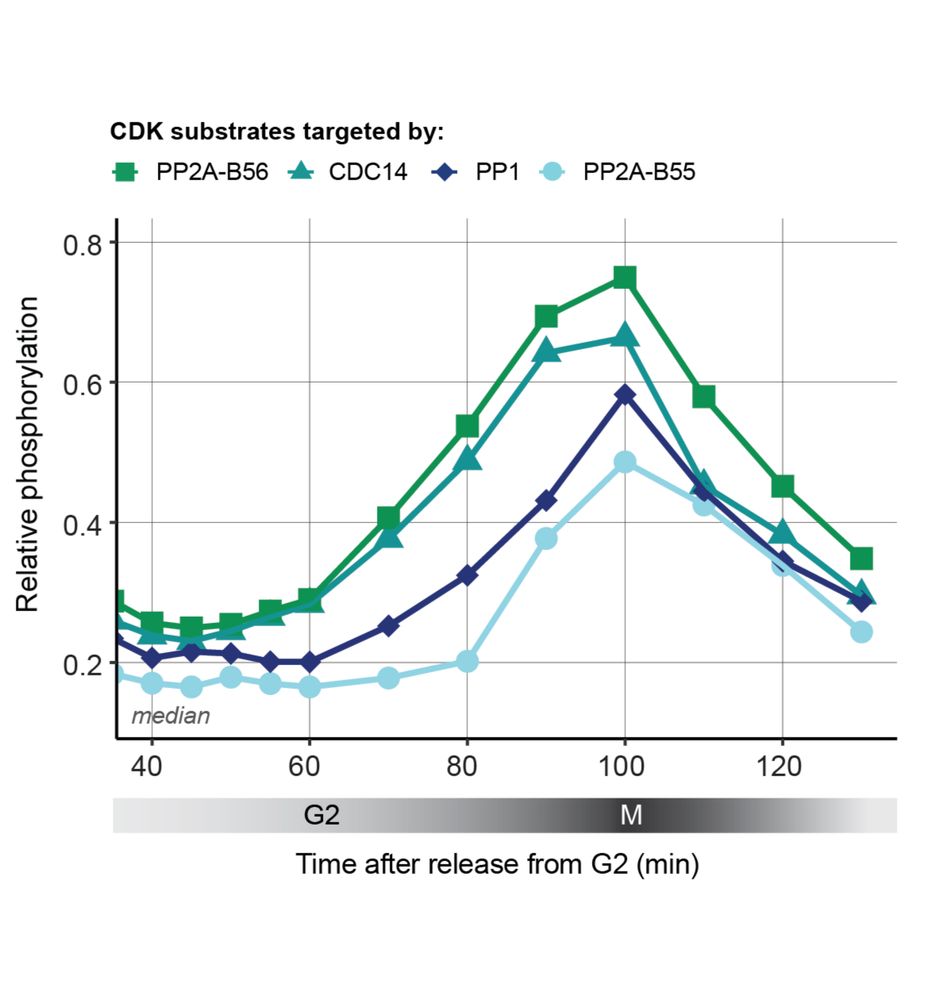
Does the division of labour between the Ppase impact the timing of CDK substrate phosphorylation? Yes! ✅
Phosphoproteomic timecourse showed that CDK substrates targeted by CDC14 and PP2A-B56 are, on average, net phosphorylated first, followed by CDK substrates targeted by PP1 and then PP2A-B55.
24.02.2025 17:09 —
👍 2
🔁 0
💬 1
📌 0
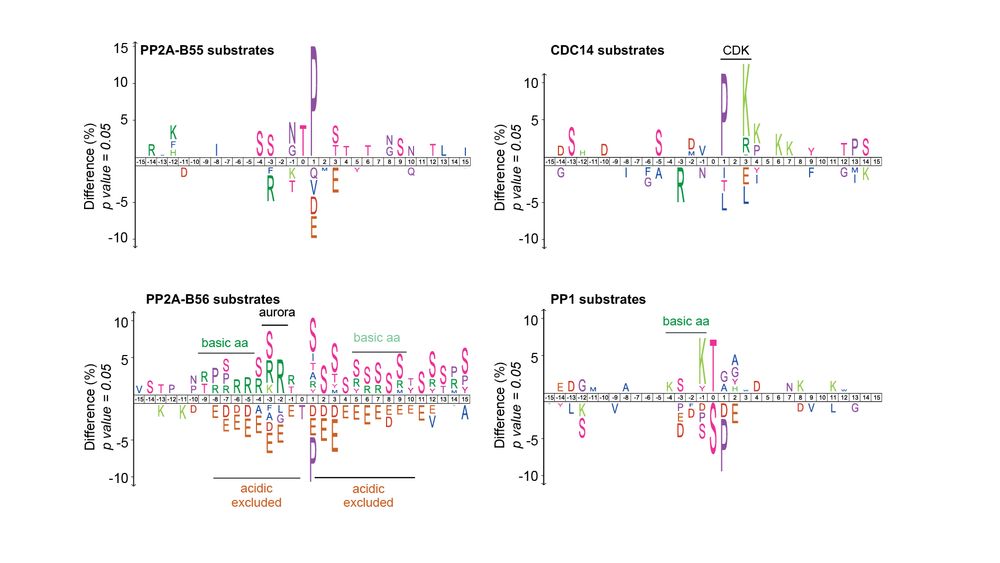
Identifying some more Ppase substrates…
We also identified 3,000+ Ppase substrates phosphorylated by other kinases (not CDK). The identified Ppase motifs were similar to those identified in other eukaryotes, indicating conservation of Ppase substrate specificity.
24.02.2025 17:09 —
👍 2
🔁 0
💬 1
📌 0
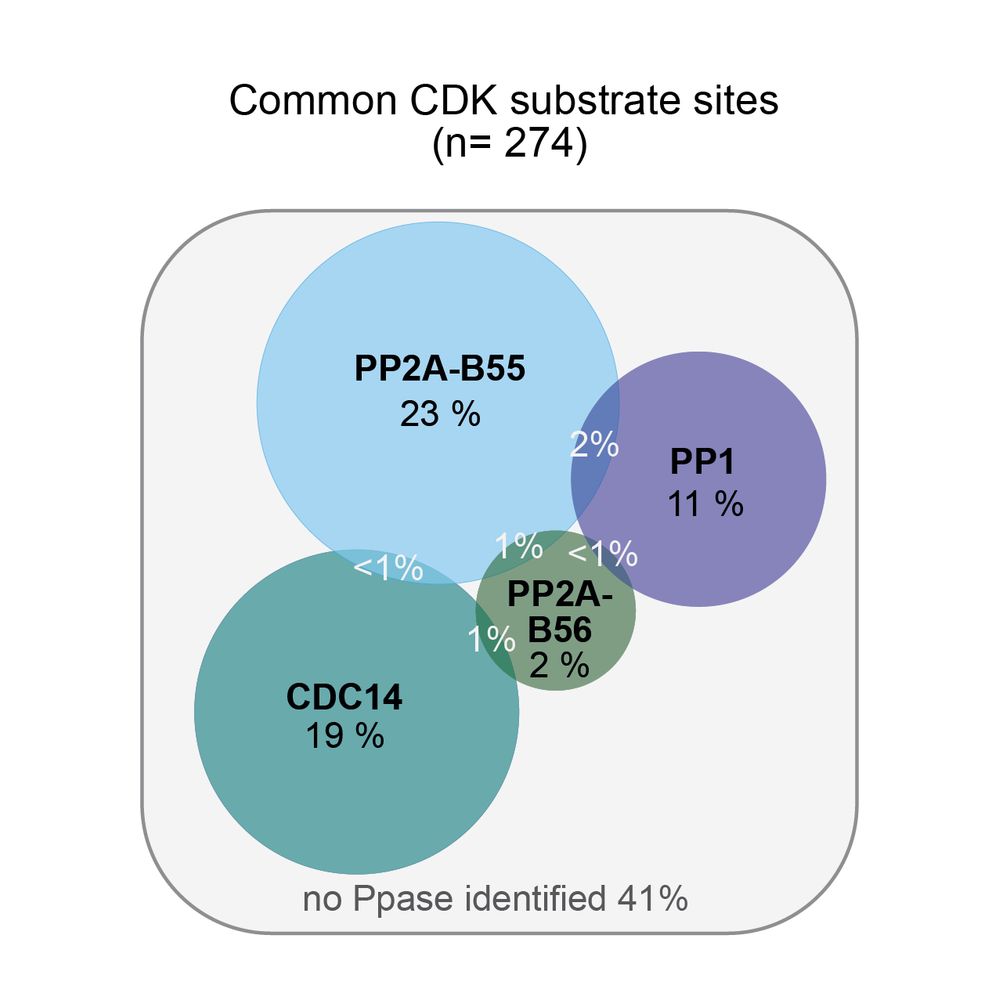
Distinct Ppase targets
• PP2A-B55, B56, CDC14 & PP1 targeted specific subgroups of CDK sites
• <5% of sites were targeted by multiple Ppases
• Ppases had different preferences for amino acids
24.02.2025 17:09 —
👍 2
🔁 0
💬 1
📌 0
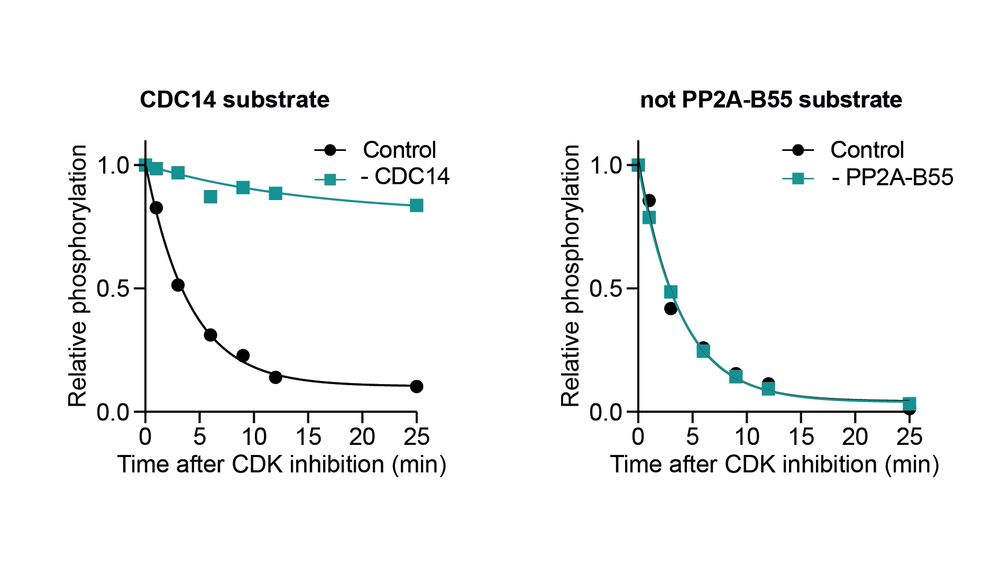
🔎 Identifying Ppase substrates
We used phosphoproteomics: inhibiting CDK with & without Ppase and tracking dephosphorylation. We classified a site as a Ppase substrate if it was dephosphorylated substantially slower in the absence of the Ppase.
24.02.2025 17:09 —
👍 2
🔁 0
💬 1
📌 0
❓Open questions
How do CDK-opposing Ppases (PP2A-B55, PP2A-B56, PP1 & CDC14) together influence the phosphorylation timing of CDK substrates?
To answer this, we first needed to identify which CDK substrate is targeted by which Ppase.
24.02.2025 17:09 —
👍 1
🔁 0
💬 1
📌 0
🔄 CDK-opposing Ppases
• Important at mitotic exit to dephosphorylate CDK substrates
• Ppases are also active during interphase ➡️ could affect phosphorylation timing
• But for the majority of CDK substrate sites, it is not known which Ppase targets it
24.02.2025 17:09 —
👍 1
🔁 0
💬 1
📌 0
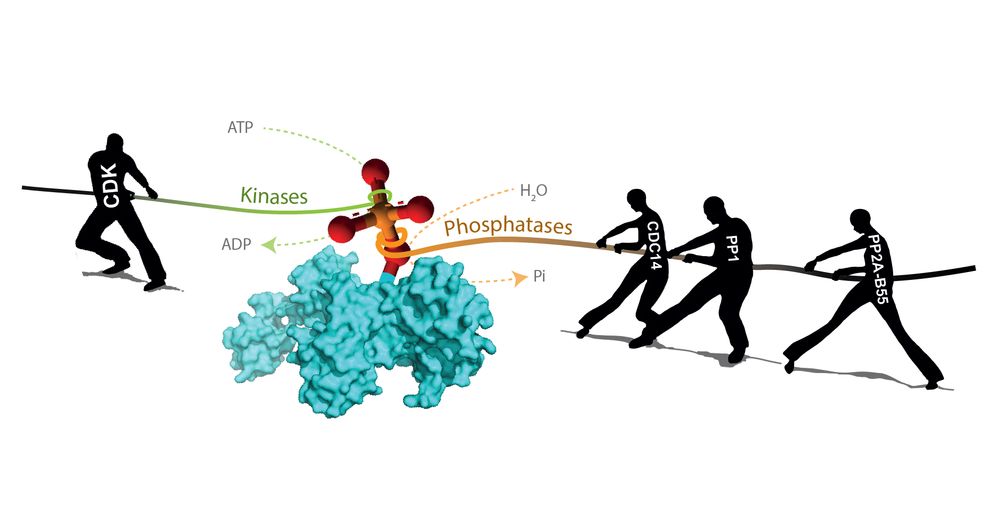
⏰ The cell cycle is driven by reversible phosphorylations
Hundreds of proteins need to be phosphorylated at the right time by cyclin-dependent kinases (CDKs) to order cell cycle events. These phosphorylations are opposed by phosphatases (Ppases), such as PP2A-B55, PP2A-B56, PP1 & CDC14.
24.02.2025 17:09 —
👍 2
🔁 0
💬 1
📌 0
🚨New preprint🚨
How do phosphatases affect CDK substrate phosphorylation timing during the cell cycle?
We showed that substrates dephosphorylated by different Ppases are net phosphorylated at different times.
📖 Read here: doi.org/10.1101/2025... & follow along for some key insights 🧵
24.02.2025 17:09 —
👍 27
🔁 8
💬 1
📌 2

Home
We’ve got loads of science experiments, quizzes and crafts for kids and families. Get exploring and see what you discover on Crick Kids.
🧪 Got a curious 7–11 year-old at home? 🔍 Whether they’re wondering how yeast grows or want to craft some red blood cells, the new Crick Kids website has them covered! 🎉
Discover fun science activities, hands-on experiments, and lots of quizzes 🧬👩🔬🔬
👉 Check it out here: kids.crick.ac.uk
02.01.2025 17:48 —
👍 6
🔁 4
💬 0
📌 0

Conference announcement
Join us in Liverpool to celebrate our 100th anniversary
Biologists @ 100
24-27 March 2025, ACC Liverpool, UK
biologists.com/100-years/conference
Register now
Extended abstract deadline:
17 January 2025
#biologists100
🧪The deadline to submit an abstract for oral and poster presentations at our Biologists @ 100 conference has been extended to Friday 17 January 2025. View the abstract topics and submit yours at 100yearsconference.biologists.com/abstracts/ #biologists100
19.12.2024 15:22 —
👍 36
🔁 29
💬 0
📌 3
















