Video snippet courtesy of Belfast legend David Holmes on Instagram...
22.02.2025 20:04 — 👍 14513 🔁 4069 💬 198 📌 178
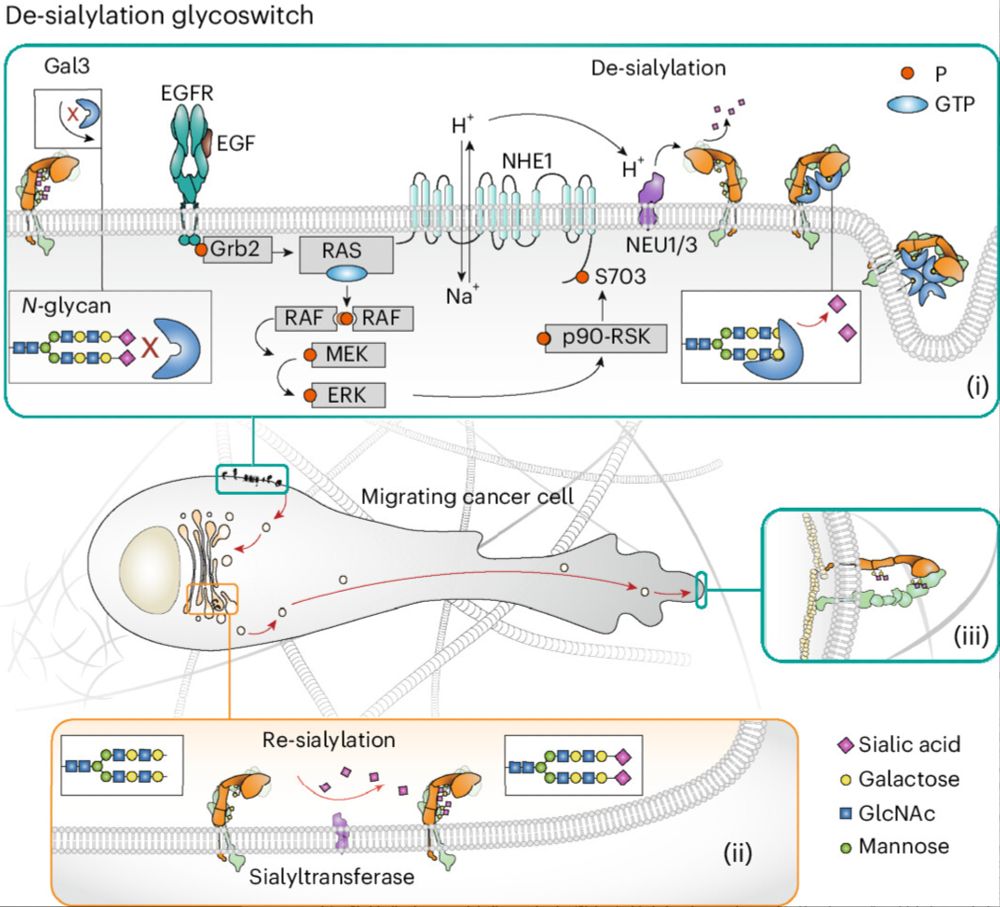
"Growth factor-triggered de-sialylation
controls glycolipid-lectin-driven endocytosis"
An excellent collaboration between the US, India, Denmark, UK, Netherlands and France.
Growth factors trigger receptor desialylation at the plasma membrane, galectin binding and subsequent endocytosis.
=> A proton pump driven glycoswitch @naturecellbiology.bsky.social :
rdcu.be/eaLM9
#glycotime, @institutcurie.bsky.social, @cnrs.fr, @inserm.fr, #NIH, #CellBiol, #Cancer
22.02.2025 22:31 — 👍 16 🔁 3 💬 2 📌 1
Happy to contribute to the analysis of β1integrin endocytosis using @the.3i.social LLSM, now in @naturecellbiology.bsky.social
rdcu.be/eaLM9
L. Leconte @christianwunder.bsky.social @aforrester.bsky.social @ewanmacdonald.bsky.social @institutcurie.bsky.social #inria @france-bioimaging.bsky.social
22.02.2025 09:01 — 👍 13 🔁 6 💬 0 📌 2

SAVE THE DATE 🗓️Ubiquitin & Friends Symposium: 8-9 May 2025 in Vienna!
Registration will open🔜 Keep an eye out for the announcement 👀
✨guest speaker lineup👇🧵 +talks from abstracts! Great chance for #ECRs to meet with peers & experts in a friendly setting!
🔗 www.protein-degradation.org/symposium/
29.11.2024 14:18 — 👍 60 🔁 28 💬 1 📌 4
We said hello to our new followers last week, but there's many more of you now! Welcome! We've created a starter pack for you, which is a work in progress.
We'd also love to hear from you about your work we've published or what kind of things you want us to post.
12.11.2024 15:43 — 👍 99 🔁 42 💬 16 📌 6
More very cool research from the Hegde Lab! Looking forward to diving in :)
🔗to the original paper out @science.org here: www.science.org/doi/10.1126/...
29.11.2024 09:55 — 👍 13 🔁 2 💬 0 📌 0





