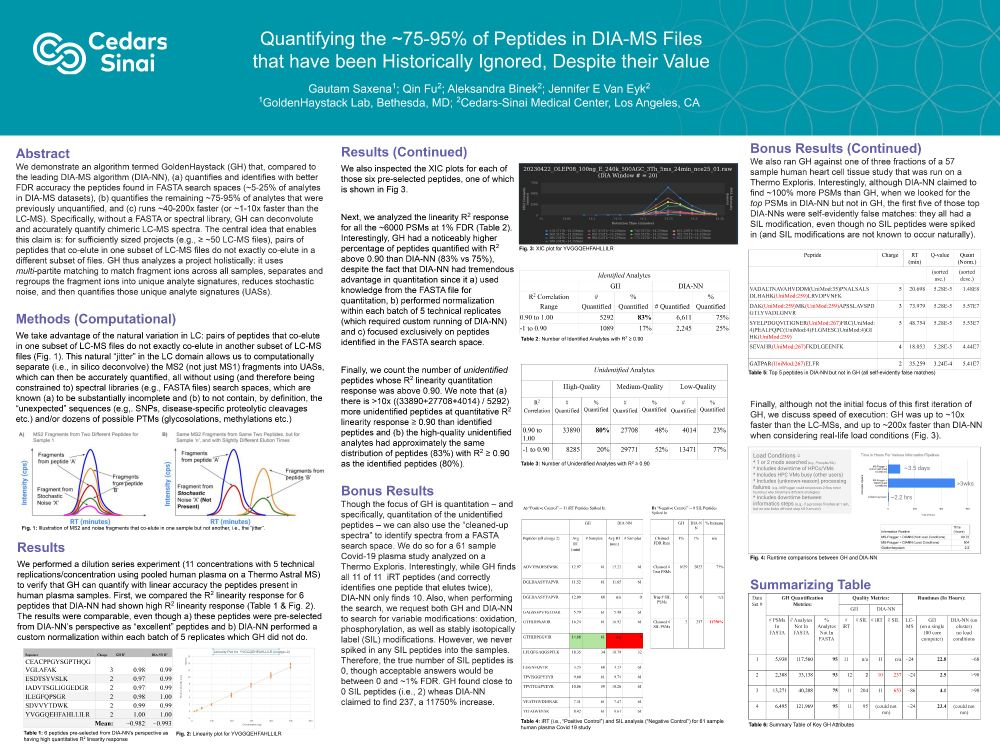
ASMS 2025 Poster in Baltimore for GoldenHaystack Lab
For those who missed me at ASMS last week in Baltimore, here's my ASMS 2025 poster. That said, we've done way more in recent months that is not disclosed in the poster, and would love, love, love to talk & collaborate. So, please do reach out to me if your lab may be interested.
10.06.2025 13:53 — 👍 0 🔁 0 💬 0 📌 0

GoldenHaystack Lab
GoldenHaystack Lab is a computationally-focused biomarker discovery lab for DIA-MS datasets that collaborates with both LC-MS instrument labs and medical research labs.
I'm the developer behind GoldenHaystack. Yes, I second Jenny -- please try it! www.goldenhaystack.org Pls email me directly (gsaxena@goldenhaystack.org) -- it should have no trouble with 1000s of DIA-MS files.
14.02.2025 13:08 — 👍 0 🔁 0 💬 0 📌 0

Quantifying the ∼75-95% of Peptides in DIA-MS Datasets that were not Previously Quantified
We demonstrate an algorithm termed GoldenHaystack (GH) that, compared to the leading DIA-MS algorithm, (a) quantifies and identifies with better FDR accuracy the peptides found in FASTA search spaces ...
For over a decade, discovery MS proteomics algorithms only measured the ~5-25% of peptides that we users "knew about", while completely ignoring the remaining (and moreover invaluable) ~75-95% that was in the MS. A ~week ago, we published a preprint showing how we can go after that ~75-95%.
01.02.2025 18:37 — 👍 3 🔁 1 💬 0 📌 0
this ~75-95% set exclusively contains the unpredicted peptide sequences & unexpected PTMs that can be invaluable to your lab (e.g., please see reference to “Azheimer study” in first paragraph of page 3 of preprint and our website). We’re seeking labs that like to collaborate. (2/2)
23.01.2025 13:32 — 👍 1 🔁 0 💬 1 📌 0

Quantifying the ~75-95% of Peptides in DIA-MS Datasets that were not Previously Quantified
We demonstrate an algorithm termed GoldenHaystack (GH) that, compared to the leading DIA-MS algorithm, (a) quantifies and identifies with better FDR accuracy the peptides found in FASTA search spaces ...
Introducing GoldenHaystack (GH) algorithm & lab (link card for *updated* preprint below; website = www.goldenhaystack.org): Labs can now quantify the ~75-95% of peptides in DIA MS files that existing algorithms do not quantify, an up to ~2000% increase in quantifiable peptides. Further, (1/2)
23.01.2025 13:31 — 👍 3 🔁 2 💬 1 📌 0
Of those 4 limiting areas, do you have a super rough sense of how much each of those areas consumed in the total 4 week time frame?
11.01.2025 21:02 — 👍 0 🔁 0 💬 1 📌 0
Thanks for reading our paper, Vadim! I emailed you directly to set up a possible meet to interactively discuss the value of using SIL-search (which are by definition false matches, unlike using sequences from entrapment dbs etc.) Thanks in advance, Vadim. -Regards, Gautam.
19.12.2024 13:13 — 👍 0 🔁 0 💬 2 📌 0




