GUANACO: A Unified Web-Based Platform for Single-Cell Multi-Omics Data Visualization https://www.biorxiv.org/content/10.1101/2025.09.18.677070v1
21.09.2025 12:47 — 👍 0 🔁 1 💬 0 📌 1
Thrilled to collaborate on cosupervising this work! GUANACO puts single-cell MultiOmics visualization in reach for bench scientists: built-in stats, no code for publication-ready figures. pls explore more functions from the link 👇Congrats to the team!
22.09.2025 06:14 — 👍 0 🔁 0 💬 0 📌 0
LinkedIn
This link will take you to a page that’s not on LinkedIn
A few weeks ago, I had to discuss about DynaTag, a smart tweak to CUT&Tag that enables single-cell transcription factor mapping with just a buffer change.
Opening a number of future avenues.
📰 Via
@GenomeWeb
: lnkd.in/djYm_sGg
#epigenetics #singlecell
10.09.2025 16:37 — 👍 13 🔁 5 💬 0 📌 0

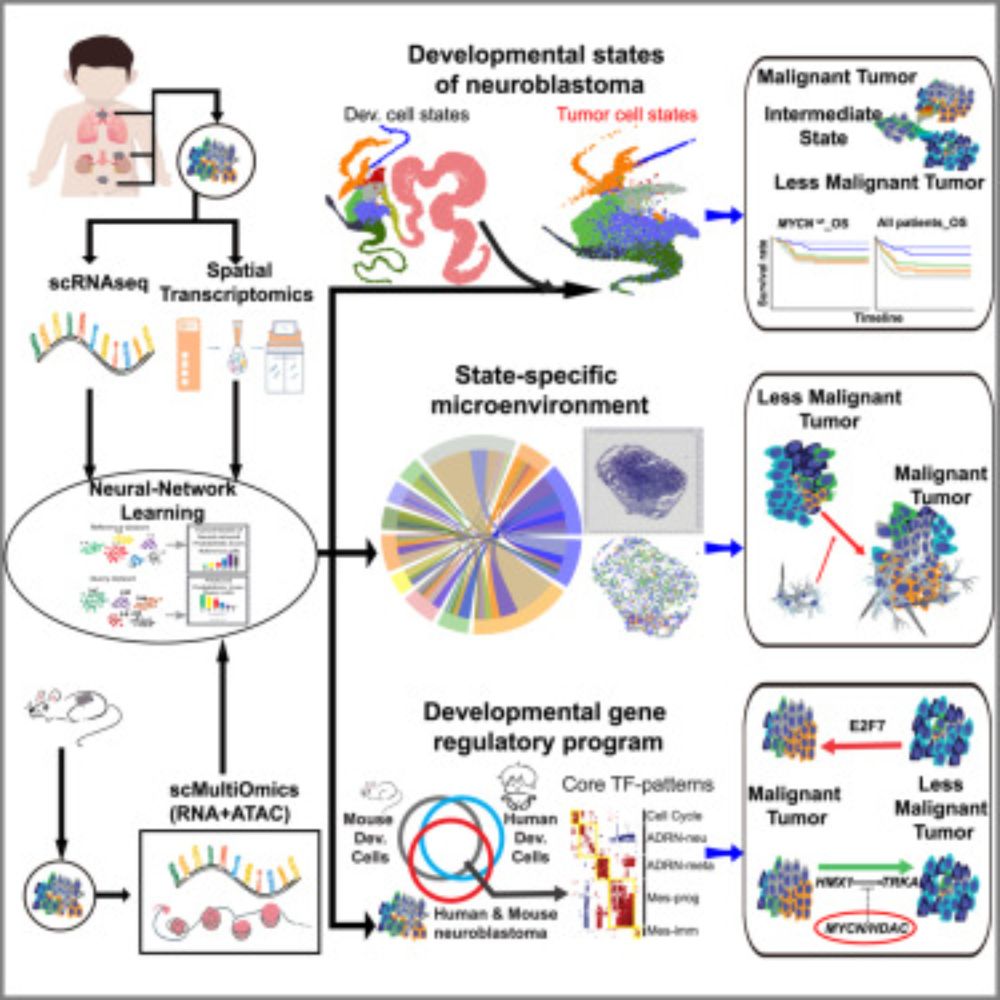
Our study on neuroblastoma developmental plasticity is the cover story in the latest issue of Dev. Cell @cp-devcell.bsky.social .The Chinese-ink Taiji (Yin–Yang) cover to echo cell-state transitions. Huge thanks to the editors, co-authors. #Neuroblastoma www.cell.com/developmenta...
10.09.2025 14:25 — 👍 3 🔁 1 💬 0 📌 0

Postdoctoral studies in single-cell and computational biology
Postdoctoral studies in single-cell and computational biology Do you want to contribute to top quality medical research? A postdoctoral position is available in the laboratory of Professor Francois
Postdoc in Computational Biology – Auditory Pathways & Sensory Disorders @LallemendLab to study how peripheral/central auditory pathways (circuit, cellular, molecular) are altered in tinnitus & age-related hearing loss. #scRNAseq, #barcode-lineage-tracing, #translational
ki.varbi.com/what:job/job...
02.06.2025 10:21 — 👍 3 🔁 3 💬 0 📌 1
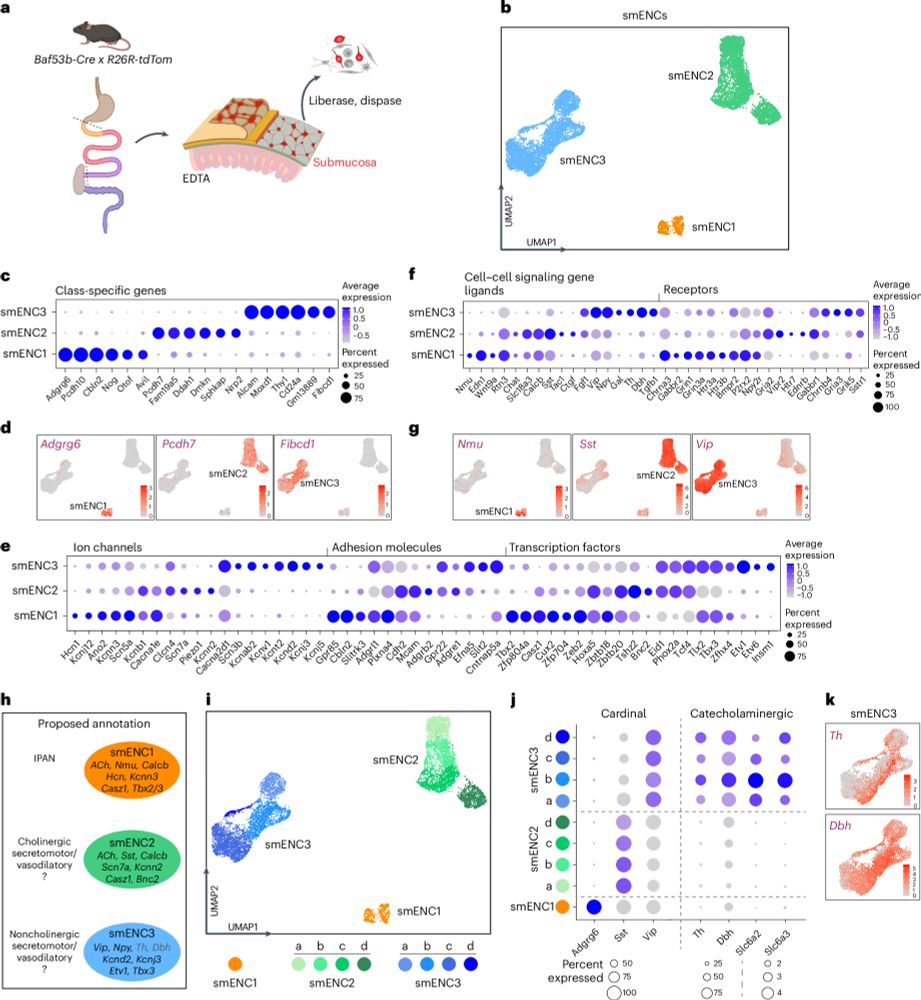
The transcriptomes, connections and development of submucosal neuron classes in the mouse small intestine
Nature Neuroscience - The neuronal composition of the intestinal submucosal plexus is incompletely understood. Here Li et al. define their neuron classes, connectome and stepwise acquisition of...
Check out our new study on the innermost nervous system of the body—the submucosal plexus. We define three neuronal classes, map their connectivity, and uncover their stepwise development via phenotypic switching. Congrats to all authors! #NatureNeuroscience #EntericNervousSystem rdcu.be/eoqXz
29.05.2025 11:28 — 👍 16 🔁 6 💬 1 📌 2
LinkedIn
This link will take you to a page that’s not on LinkedIn
Epichrom 2025 !
After 3 years, Epichrom is revived and will take place in Stockholm on the 12th of June. Great opportunity to share your research and hear about latest epigenetics/chromatin research in Sweden.
Registration is free of charge
www.bartosovic-lab.com/epichrom
07.05.2025 05:23 — 👍 4 🔁 4 💬 1 📌 0
Thank you @ki.se for sharing our @cp-devcell.bsky.social study on neuroblastoma developmental plasticity! 🙏🚀
12.05.2025 15:13 — 👍 1 🔁 0 💬 0 📌 0
Thank you, Susanne! Your help made this possible.
10.05.2025 10:52 — 👍 1 🔁 0 💬 0 📌 0
9/9🙏 Thanks to our collaborators:
@schlisio-lab.bsky.social
, Prof. Arthur Tischler (@tuftsmedicalcenter.bsky.social), Prof. Ronald de Krijger (
@themaximacenter.bsky.social
), and all the other amazing team members!
09.05.2025 22:07 — 👍 3 🔁 0 💬 0 📌 0
scCAMEL — scCAMEL 0.26b documentation
8/9 Our enhanced neural-network tool, scCAMEL, now aligns cross-species, multimodal data in a single workflow—empowering comprehensive mapping of tumor plasticity. sccamel.readthedocs.io/en/latest/
09.05.2025 22:04 — 👍 0 🔁 0 💬 1 📌 0
7/9 Spatial maps reveal clonal pockets of bridge & neural‑crest‑like cells encircled by TAMs & CAFs that sustain aggressiveness—micro‑environment is a Dev. state‑specific partner‑in‑crime.
09.05.2025 22:04 — 👍 0 🔁 0 💬 1 📌 0
6/9 Functional hits 👉 E2F7-networks fuel de‑differentiation, while HMX1 can force differentiation—unless MYCN amplification blocks it.---> Dev. state‑specific vulnerabilities = therapeutic targets.
09.05.2025 22:03 — 👍 0 🔁 0 💬 1 📌 0
5/9 🔑 Enhancer–gene regulatory networks uncover 121 core TF gene patterns organized into five principal patterns: Cell Cycle, ADRN-meta, ADRN-neu, Mes-Prog, and Mes-immune.
09.05.2025 22:03 — 👍 1 🔁 0 💬 1 📌 0
4/9 The enhancer‑primed epigenetic landscape gives neuroblastoma cells the "latent" capacity to shift between developmental programs.
09.05.2025 22:03 — 👍 0 🔁 0 💬 1 📌 0
3/9 We traced 7 NBL cell states back to the trunk neural crest lineage. A previously less-appreciated “bridge” state acts as the hub for state switching and, together with neural‑crest‑like cells, dominates high‑risk NBL.
09.05.2025 22:03 — 👍 0 🔁 0 💬 1 📌 0
2/9 In collaboration with
@patrikernfors.bsky.social
and Jian Wang’s team, we integrated single-cell and spatial transcriptomics from patient tumors and scRNA+ATAC data from mouse spontaneous models to construct a high-resolution disease atlas.
09.05.2025 22:02 — 👍 0 🔁 0 💬 1 📌 0

Join us at the Nobel Symposium
Multiple Sclerosis: Past, Present and Future Treatment
June 5th, 2025 | Jacob Berzelius lecture hall @ki.se | Stockholm, Sweden
free registration at:
jirango.com/cview/web/f0...
05.05.2025 10:48 — 👍 14 🔁 5 💬 1 📌 0
New mug, #science_doesn’t_care_what_U_believe
13.03.2025 14:42 — 👍 0 🔁 0 💬 0 📌 0
Research group leader @karolinskainst & @helsinkiuni. Lab website: https://www.chen-sysimeta-lab.com/
Reviews journal by Cell Press publishes articles covering all aspects of human diseases, diagnostics, therapeutics, and disease prevention. Posts are by the editor, Dr. Aliki Perdikari. Explore more at: https://www.cell.com/trends/molecular-medicine/home
UCSD professor of neurobiology and neurosciences
Associate Professor of Developmental and Evolutionary Genomics at Imperial College London, Department of Life Sciences. Spent a decade in Philly, #FlyEaglesFly
https://marcotrizzino.wordpress.com/
EMBO Postdoctoral fellow @makinenlab | PhD @ralfhadams | Interested in Arteries, Veins, Capillaries, Lymphatics and Food. x.com/vishalmkrishnan
Stem cell and mechanobiologist at the Max Planck Institute of Molecular Biomedicine and University of Helsinki
Neuroscience, meninges, blood-brain barrier, cake (or cookies)
http://siegenthalerlabcu.weebly.com/
MD/PhD Student
University of Wisconsin School of Medicine and Public Health
Medical Scientist Training Program, Class of 2027
Interested in pediatric oncology, myeloid immunotherapies, and narrative medicine
Group leader
@sangerinstitute.bsky.social Neural diversity, spatial transcriptomics, glia, GBM
Our group develops and applies computational approaches to study molecular variations and their phenotypic consequence. We are part of DKFZ and EMBL.
Website: https://steglelab.org/
Scientist at DKFZ and EMBL in Heidelberg, loving stats, genomics and genetics. @OliverStegle@genomic.social. For group news see @steglelab.bluesky.social
Activities of the Neurooncology Community in Heidelberg
I research pediatric brain tumors at Uppsala University 🇸🇪. Cancer genome enthusiast and northern lights seeker
Société Française de Biologie du Développement
French Society for Developmental Biology
3D Human Brain Models Lab ,Personalised models, #Microenvironment , @uniklinik_fr #WomeninScience ,Chief Organizer of http://ngi-freiburg.com, Replacement of Animals #3R
https://www.uniklinik-freiburg.de/neurochirurgie/forschung/3d-brain-models-lab
Associate Professor & Docent @umeauniversitet.bsky.social
Wallenberg Fellow in Molecular Medicine at WCMM Umeå
Postdoc @embl.org
PhD @cniostopcancer.bsky.social
3D genomics , epigenomics, gene regulation, enhancer, cancer, glioblastoma, cancer neuroscience
Physician scientist, Glioma single-cell genomics researcher, Neurosurgeon @Tokyo University, Postdoc in Suva lab @MGH/Broad
Medical oncologist and physician scientist @TLVMC; PhD graduate @TiroshLab @WeizmannInstitute; Opinions are my own