How do DNA sequence and histone composition modulate nucleosome plasticity? We investigated this by comparing the behaviour of 40 chemically different nucleosomes. Check our preprint below.
27.02.2026 09:19 — 👍 11 🔁 4 💬 0 📌 0How do DNA sequence and histone composition modulate nucleosome plasticity? We investigated this by comparing the behaviour of 40 chemically different nucleosomes. Check our preprint below.
27.02.2026 09:19 — 👍 11 🔁 4 💬 0 📌 0Really excited to share the latest work from my PhD with @giuliotesei.bsky.social and @lindorfflarsen.bsky.social!
09.02.2026 12:55 — 👍 6 🔁 4 💬 1 📌 0
Want to learn about how we use computational approaches at different scales to study biomolecular condensates?
Check out our latest review, out in Advances in Physics X
@juliamaristany.bsky.social @alinaemelianova.bsky.social
@rcollepardo.bsky.social
www.tandfonline.com/doi/epdf/10....
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
Excited to see our review now on arXiv, written together with @fpesce.bsky.social and @lindorfflarsen.bsky.social
doi.org/10.48550/arX...
New preprint is out !
Bad news : current all-atom simulations of phosphorylated IDPs are very probably wrong (and yes, this is clickbaity on purpose 😇)
Good news : we know what to blame for it, and we even have an idea of how to fix it !
Our paper on:
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
Arriën & Giulio's paper on
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
AlphaFold is amazing but gives you static structures 🧊
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
Led by @sobuelow.bsky.social & @giuliotesei.bsky.social we put together a full overview of the CALVADOS software and applications.
Give it a read: doi.org/10.48550/arX...
Give it a try: github.com/KULL-Centre/...
Thanks to @lindorfflarsen.bsky.social and all authors for this wonderful project on predicting IDR phase separation from sequence!
Check out the published version (including added exp. data from @tanjamittag.bsky.social) and feel free to try out our webserver.
Very happy to share our next extension to the CALVADOS protein force field:
If you want to explore the changes in global dimensions of a disordered protein upon phosphorylation: give it a read and a try!
Big thank you to @giuliotesei.bsky.social, @lindorfflarsen.bsky.social and Gustav S. Hedemark

Big thank you to Giulio Tesei and @lindorfflarsen.bsky.social for their guidance.
The preprint can be found here: www.biorxiv.org/content/10.1...
The code and data are available here: github.com/KULL-Centre/...
Excited to share our PEG model for disordered proteins in CALVADOS!
If you are interested in exploring the effects of a crowder on the global dimensions of an IDP or want to explore the phase separation behaviour of a more weakly PS-prone IDP, have a look at our preprint and give it a try.

Figure from manuscript showing epresentative snapshots from slab simulations with 100 chains A1 (dark blue) with w/v-percentages of 0%, 5% (100:127) and 10% (100:254) PEG8000 (orange chains, 181 monomers) illustrating how PEG both lowers the 𝐶sat with increasing concentrations and also increases the effective protein concentration in the condensate (𝐶con).
Preprint submitted 🇺🇦
01.03.2025 22:22 — 👍 19 🔁 1 💬 0 📌 0
Figure from the paper that illustrates the approach of probing the transition state for amyloid growth by experiments and simulations
How do proteins mis-fold?
Paper led by Jacob Aunstrup from Alex Büll’s lab with MD simulations by Abigail Barclay, and key contributions from several others. We combined measurements of Φ-values with MD simulations to study the transition state for amyloid fibril growth
doi.org/10.1038/s415...
Check out @rasmusnorrild.bsky.social's work with Alex Buell and Joe Rogers developing and using Condensate Partitioning by mRNA-Display to probe phase separation of ~100.000 sequences, and @sobuelow.bsky.social's simulations to support and analyse the experiments
www.biorxiv.org/content/10.1...
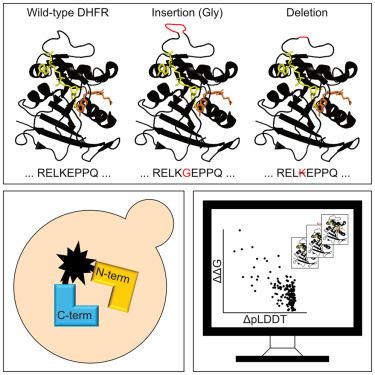
In our new paper we characterize the effects of indel variants on protein folding and stability using a new yeast-based protein folding sensor. The work was led by Sven Larsen-Ledet and supported by the @novo-nordisk.bsky.social.
Link to paper: www.cell.com/structure/fu...
Meet the CALVADOS RNA model
Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪

Flyer for the course Keeping up with the interactome - How IDPs work including a list of speakers: Professor Tanja Mittag, St Jude Children’s Hospital, Memphis, US Professor Kresten Lindorff-Larsen, BIO, SCIENCE, UCPH Professor Ylva Ivarsson, Uppsala University, Sweden Dr. Michael Wehr, Systasy Bioscience, Munich, Germany Professor Zsuzsanna Dostztányi, Eötvös Loránd University, Hungary Professor Kristian Strømgaard, HEALTH, UCPH Professor Stefano Gianni, Sprienza University of Rome, Italy Professor Per Jemth, Uppsala University, Sweden Dr. Franziska Schöppe, Novo Nordisk A/S, Denmark Professor Benjamin Schuler, University of Zurich, Switzerland Professor Alfonse De Simone, University of Naples, Italy Dr. Malene Ringkjøbing Jensen, CNRS, Grenoble, France Alleyn Plowright, Chief Scientific Officer, Pangea Bio, London UK Professor Birthe B. Kragelund, BIO, SCIENCE, UCPH
If you are a PhD student and like protein disorder (or want to learn more), Birthe Kragelund @bbkrage.bsky.social and Kristian Strømgaard are organizing a PhD course on How IDPs work 🧶🧬🧪 Note the cost if you are not at a Danish university.
Details: phdcourses.ku.dk/DetailKursus...
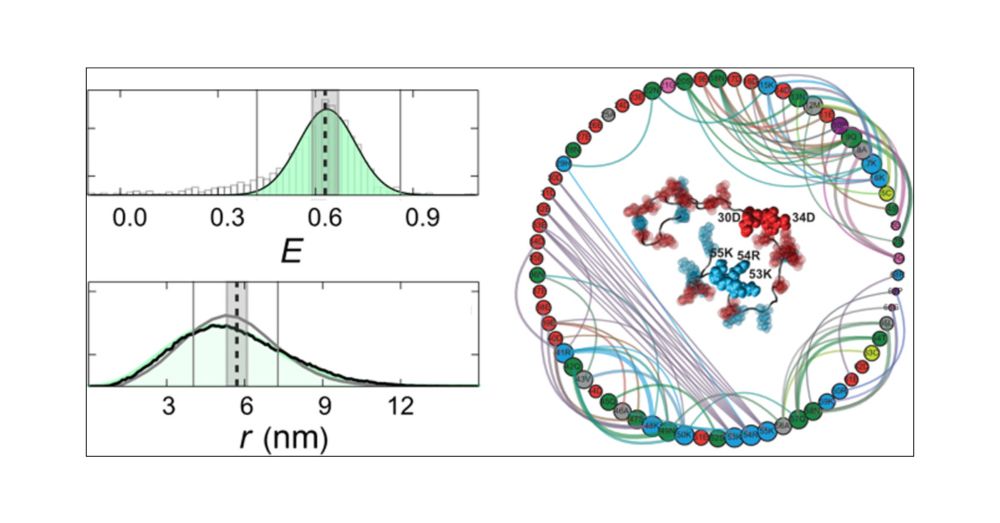
New collaborative work in collaboration with the labs of Robert Best & Tanja Mittag driven by Ben Schuler.
Identifying Sequence Effects on Chain Dimensions of Disordered Proteins by Integrating Experiments and Simulations.
pubs.acs.org/doi/10.1021/...