We are excited to share our new preprint demonstrating that nucleic acid interactions with SUZ12 constrain PRC2 activity, establishing a kinetic buffer essential for targeted gene silencing and revealing vulnerabilities in diffuse midline gliomas.
www.biorxiv.org/content/10.1...
23.07.2025 23:38 — 👍 38 🔁 17 💬 1 📌 0
Thank you for your lovely preview of our work! 😄
07.06.2025 23:55 — 👍 1 🔁 0 💬 1 📌 0
I find unstructured regions in proteins enigmatic and very interesting.
06.06.2025 20:51 — 👍 5 🔁 1 💬 1 📌 0
Thank you Elena! Congrats on your recent Mol Cell paper too 🥳 Hope you’re well!
21.05.2025 18:23 — 👍 1 🔁 0 💬 0 📌 0
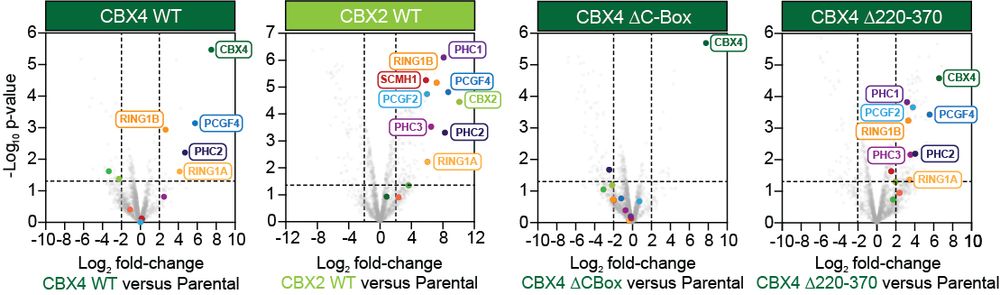
IP-mass spec revealed CBX4 preferentially forms a distinct complex with essential PCGF4. CBX2 lacks this specificity. Deleting the key disordered region shifts complex composition – the PCGF4 preference is lost, mimicking CBX2. This region defines CBX4’s unique function. (10/11)
21.05.2025 16:12 — 👍 0 🔁 0 💬 1 📌 0
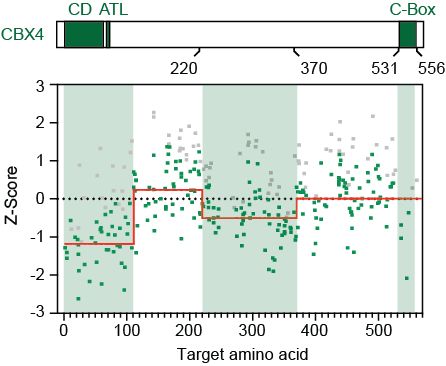
But what makes CBX4 so unique? We used a CRISPR tiling screen to pinpoint key domains. Result: the chromodomain (H3K27me3 binding) and a section of the central disordered region are essential for function. The chromodomain makes sense - what about that disordered part? (9/11)
21.05.2025 16:12 — 👍 0 🔁 0 💬 1 📌 0
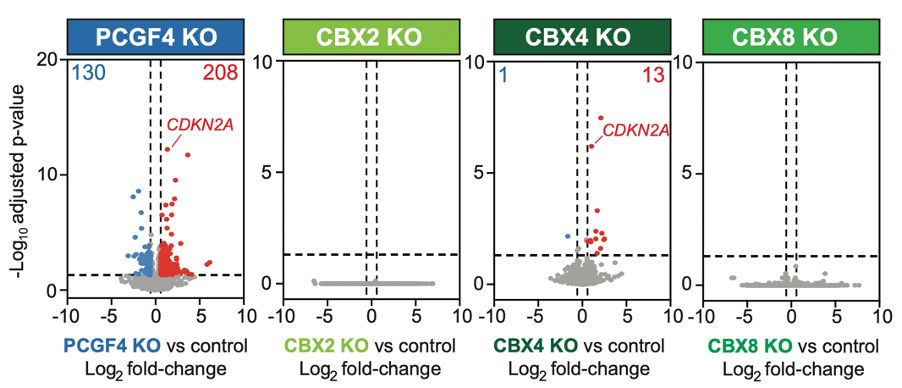
CUT&RUN-Rx says no - all forms of cPRC1 are displaced equally by PRC2i. Still, part of our hypothesis held: CBX4-cPRC1 drives repression of key genes, like p16-INK4A. KO of CBX4 derepressed genes, while KO of abundant CBX2/CBX8 had no significant impact on gene expression! (8/11)
21.05.2025 16:12 — 👍 0 🔁 0 💬 1 📌 0
H3K27me3 retention and CBX4-cPRC1 accumulation at Polycomb target genes correlates with stronger gene repression in DMG. PRC2 inhibition derepresses target genes and DMG is sensitive to this. We thought: could PRC2 inhibition work by selectively displacing CBX4-cPRC1? (7/11)
21.05.2025 16:12 — 👍 0 🔁 0 💬 1 📌 0
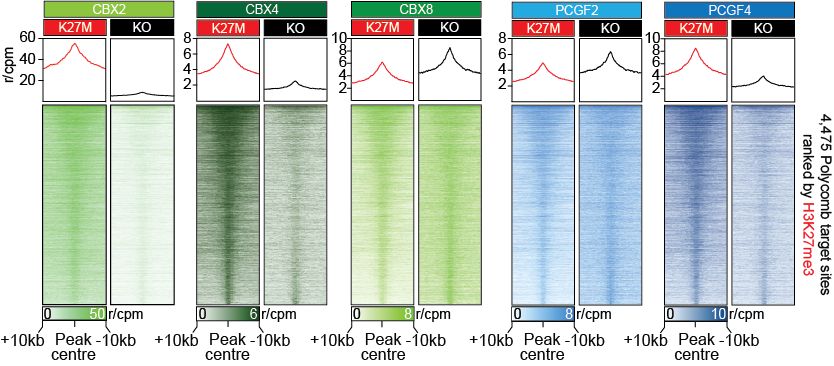
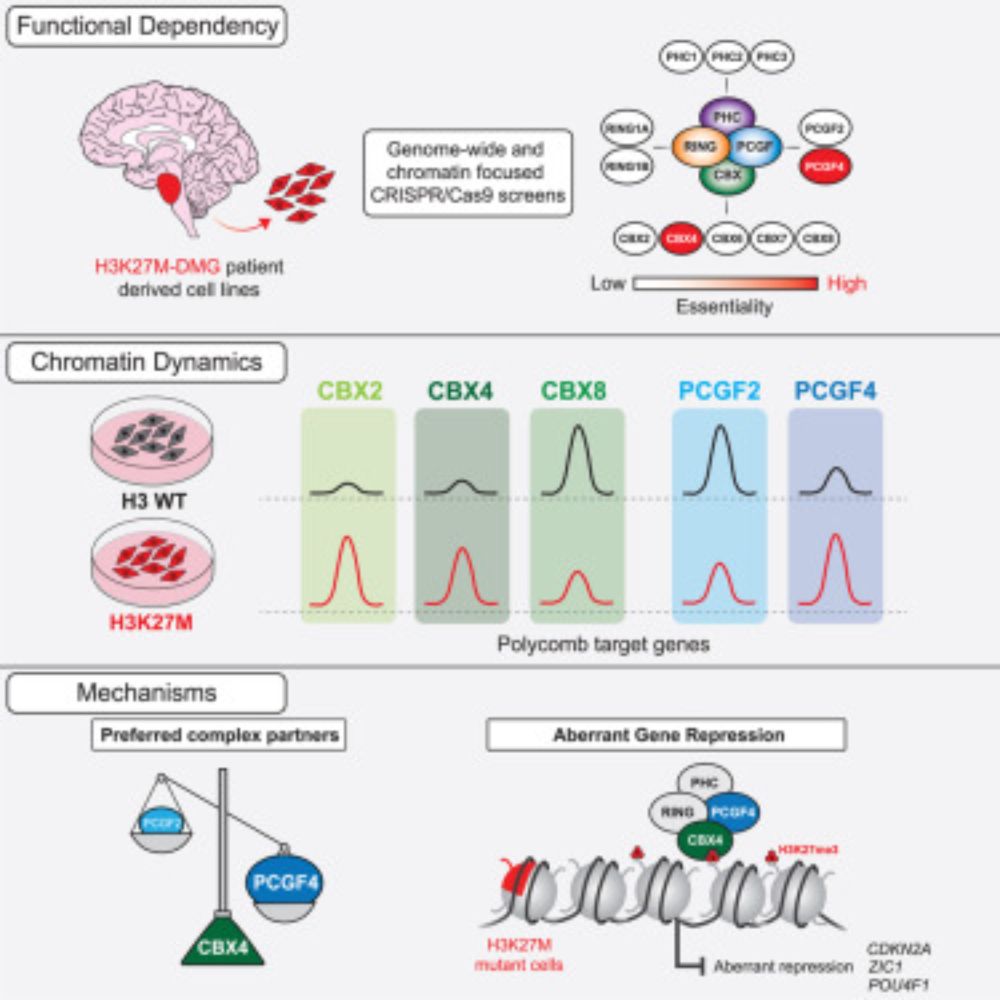
To dig deeper, we used CUT&RUN-Rx to map cPRC1 complexes in isogenic DMG cells with or without H3K27M. Strikingly, H3K27M rewires cPRC1 binding, driving CBX2/CBX4 accumulation and displacing CBX8 from CGIs at Polycomb targets genes. (6/11)
21.05.2025 16:12 — 👍 0 🔁 0 💬 1 📌 0
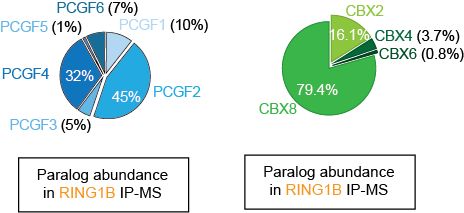
This was surprising because paralogs like PCGF2, CBX2 and CBX8 are also expressed!
Mass spec revealed CBX4 is <5% of the CBX pool in DMG cells, while non-essential CBX2 and CBX8 are far more abundant. Yet, it’s this tiny CBX4-cPRC1 minority that’s functionally essential! (5/11)
21.05.2025 16:12 — 👍 0 🔁 0 💬 1 📌 0
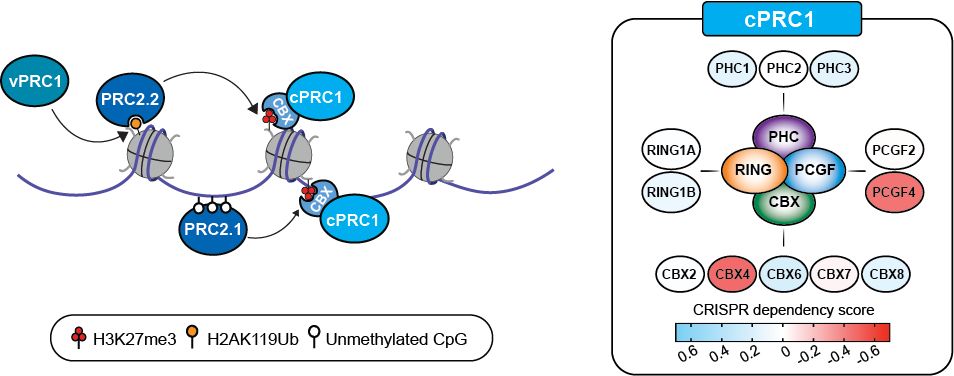
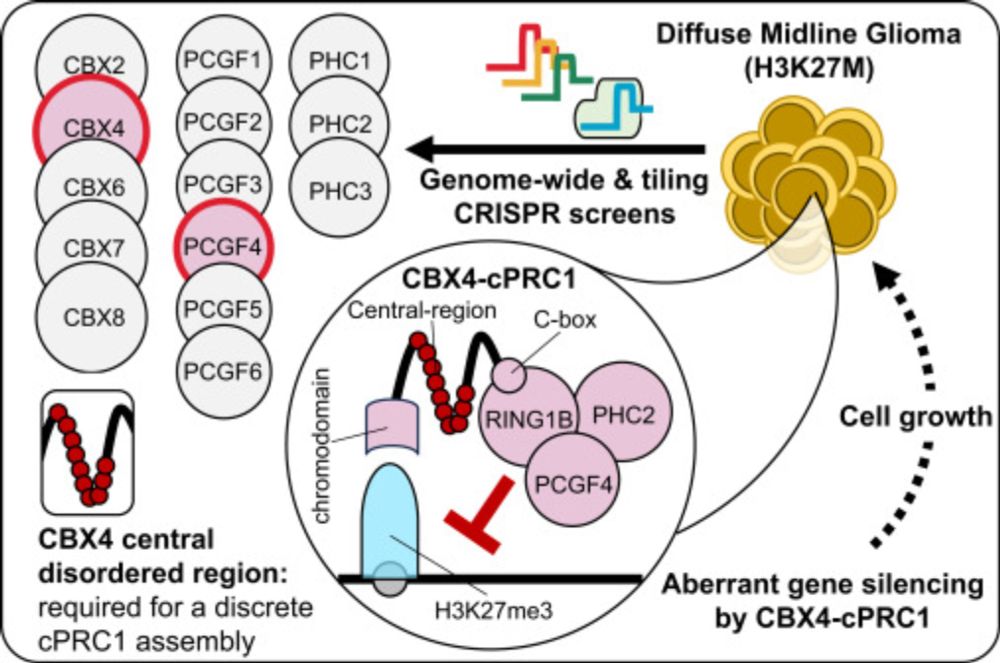
What about canonical PRC1? cPRC1 complexes bind H3K27me3 via the CBX subunit to repress gene expression. Our CRISPR-Cas9 screens in patient-derived DMG lines showed that only 2 specific cPRC1 components - CBX4 and PCGF4 (aka BMI1) - are functionally essential in DMG. (4/11)
21.05.2025 16:12 — 👍 0 🔁 0 💬 1 📌 0
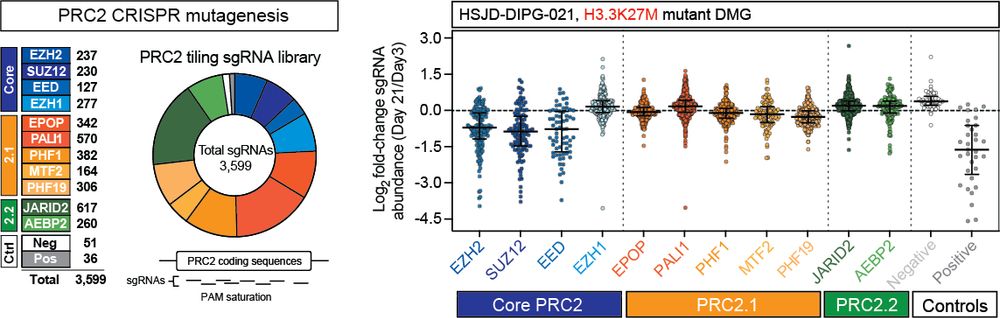
It was known that while H3K27M inhibits PRC2, residual H3K27me3 at CGIs is crucial for repressing oncogenic programs in DMG. We built on this by showing that while all PRC2 component combinations exist in DMG cells, only the catalytic core is essential for disease biology. (3/11)
21.05.2025 16:12 — 👍 0 🔁 0 💬 1 📌 0
First, this was a collaborative effort that would not have been possible without super talented co-first author @dairegannon.bsky.social, co-senior authors Dr Richard Phillips, @adrianbracken.bsky.social & @gerrybrien.bsky.social. Thanks to many other international collaborators too! (2/11)
21.05.2025 16:12 — 👍 0 🔁 0 💬 1 📌 0
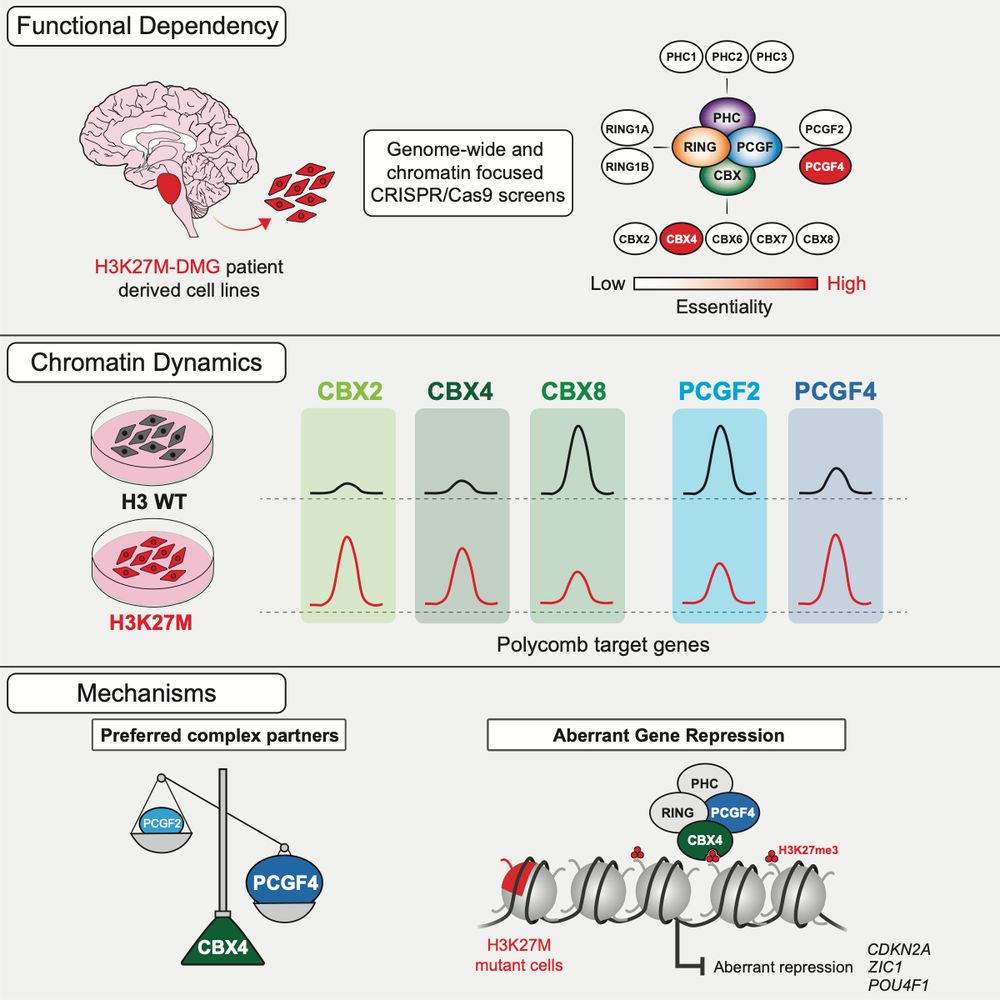
Excited to share our new paper out today in @cp-molcell.bsky.social! We show that the H3K27M oncohistone rewires cPRC1, creating a unique dependency on CBX4/PCGF4-containing complexes, and also reveal a previously unknown function of CBX4. Highlights below (1/11).
21.05.2025 16:12 — 👍 24 🔁 10 💬 5 📌 2
A career network featuring science jobs in academia and industry.
Visit our platform at www.science.hr
Engineering cells to understand their decisions
ESPOD Fellow @ebi.embl.org & @sangerinstitute.bsky.social
(Saez-Rodriguez & Parts)
lingering scientist @crick.ac.uk (Briscoe)
Scientist | Collaborative Leadership Consultant for STEM Leaders and Organizations at BridgUs Lab
Retired researcher in Developmental Genetics, Evo-Devo. Drosophila. Polycomb Group. (PRC genes). Finder of "polyhomeotic", Nuclear transplantation in the egg and epigenesis of Drosophila. Start at CSIC (Antonio Garcia Bellido) --> CNRS (M. Zalokar) etc.
PhD Student in the Long Lab 👩🏼🔬🧬 @IGC University of Edinburgh
Co-Creating Ireland's Public Involvement in Open Research Roadmap
ENGAGED is building a national roadmap to shape public involvement in open research in Ireland. We believe that research can and does play an important role in tackling societal challenges.
Award-winning Science events for kids. Founded Ireland☘️Scientist & Mum, speaker United Nations 🇺🇳 Hands-on, STEM fun to kids Ireland 🇮🇪 UK 🇬🇧Canada 🇨🇦 Join us as Franchise Owners! Licences for universities and corporations junioreinsteinsscienceclub.com
Childhood Cancer Epigenetics and Drug Discovery
Assistant Prof. at Case Western Reserve University
gryderlab.com
Chromatin and transcription aficionado. Postdoc in Bickmore lab.
Enthusiastic researcher in structural biology, genome stability, and epigenome integrity
Bioinformatics Group leader at Institute of Integrative Cell Biology (I2BC), Paris-Saclay, France. Protein addiction, interaction networks, molecular evolution & engineering, genome integrity
The mission of the Center for Physical Genomics and Engineering at Northwestern University’s McCormick School of Engineering is to create new strategies for the treatment of disease and the reversible manipulation of living systems
Assistant Professor @Cornell University
- Germline gene regulation, gene silencing, chromatin
- jongminkimlab.org
Biochemist, PhD, @LCBM, EPFL🇨🇭
PTMs | ubiquitin | single-molecule enzymology🔬| chromatin | PRC1 mediated gene regulation 🧬
Laboratory of Biophysical Chemistry of Macromolecules (LCBM) headed by @beatfierz.bsky.social at EPFL | Run by LCBM members
Scientist interested in heterochromatin formation and transcription regulation, studying transposons in @rberrens.bsky.social lab @uniofoxford Former Brockdorff lab member.
Assoc Prof at EPFL, Chemical Biology and Biophysics, working in protein chemistry, peptides, epigenetics, chromatin, microtubules
Scientist in North Carolina studying chromatin remodelers and transcription factors. @fnucleosome.bsky.social coorganizer
ORCID: 0000-0002-6256-144X
Views and posts are all my own
Epigenetics enthusiast and instructor in Emily Bernstein's lab @IcahnMountSinai | former @MSCActions PhD student in Maite Huarte's lab
Mathematical/physical/computational biologist. Postdoc @johninnescentre.bsky.social. Mostly about Science, occasionally something else. Opinions my own, even when wrong.