and our extensive exploration of scoring isogenic CRISPR screens @donnellycentre.bsky.social @sickkidsto.bsky.social www.biorxiv.org/content/10.1...
04.02.2026 09:09 — 👍 1 🔁 0 💬 0 📌 0
This chemical-genetic interaction map complements our efforts to map genetic interactions: www.biorxiv.org/content/10.1...
04.02.2026 09:05 — 👍 1 🔁 0 💬 1 📌 0
And more isogenic CRISPR screens coming out of Toronto @sickkidsto.bsky.social, this time by Mike Tyers and team - congrats! www.biorxiv.org/content/10.6...
04.02.2026 09:02 — 👍 1 🔁 1 💬 1 📌 0
Great new work from @colmr.bsky.social predicts a cancer dependency map for paralogs: doi.org/10.64898/202....
23.01.2026 11:33 — 👍 1 🔁 1 💬 1 📌 0
Perhaps one of the most anticipated data in my recent memory- amazing talk by Carmen from @sangerinstitute.bsky.social at @eurocancerdepmap.bsky.social
20.11.2025 17:16 — 👍 3 🔁 0 💬 0 📌 0
Truly inspiring talk by @icrlordlab.bsky.social at @eurocancerdepmap.bsky.social
20.11.2025 17:12 — 👍 3 🔁 0 💬 0 📌 0
Thank you @ioriolab.bsky.social for having me. It was great to talk science. @humantechnopole.bsky.social is truly a magnificent place to do research.
21.10.2025 18:05 — 👍 1 🔁 0 💬 0 📌 0
Thank you @colmr.bsky.social for the shoutout! It was quite a journey. If it is only nearly as useful as the @depmap.org we are happy. In any case, it complements this data ...as expected!?
20.08.2025 12:54 — 👍 1 🔁 0 💬 1 📌 0
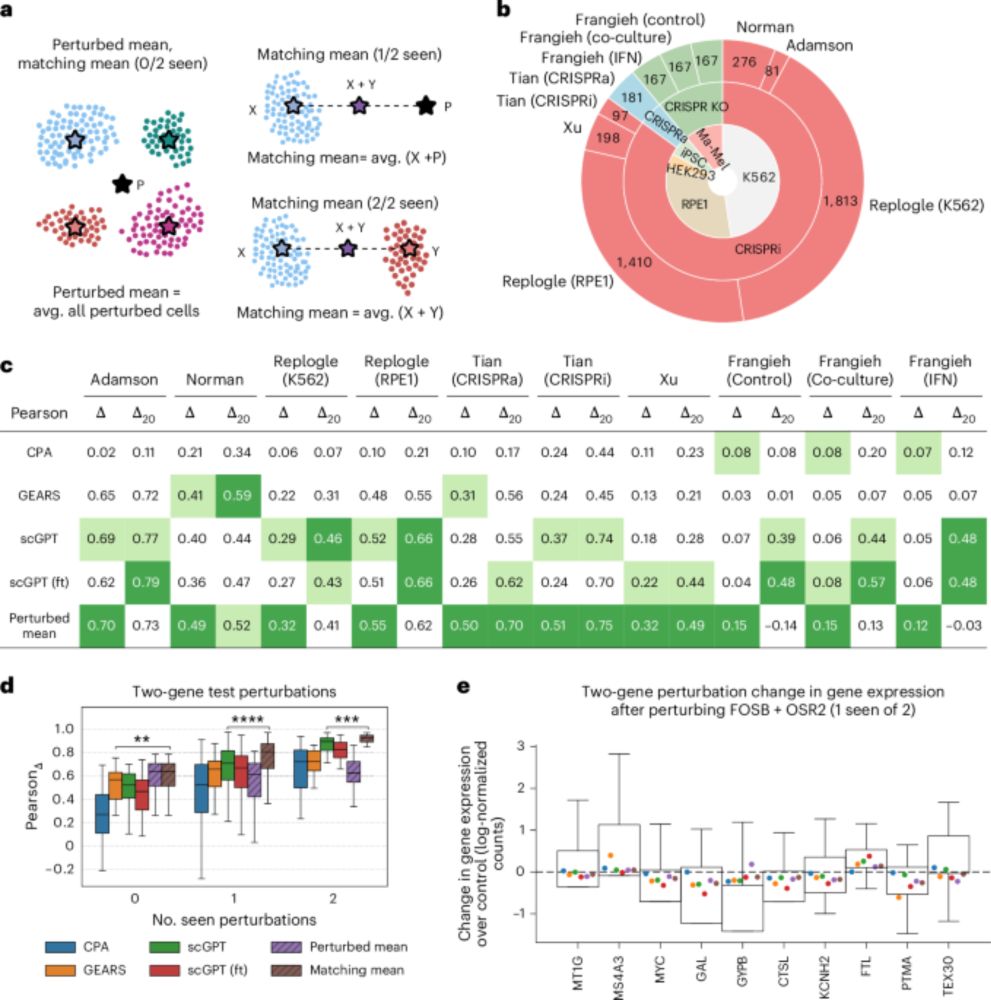
One thing that really bothers me with the new "virtual cell" terminology is that it is currently largely focused on a very narrow definition of models that can predict effects of trans perturbations (gene dosage, drugs etc) on gene expression. 1/
28.06.2025 10:38 — 👍 105 🔁 30 💬 1 📌 0
agree.
17.06.2025 13:12 — 👍 1 🔁 0 💬 0 📌 0

Jason Moffat at #VariantEffect25
22.05.2025 15:54 — 👍 3 🔁 2 💬 0 📌 0

Have you discovered the Night Science Podcast yet?
@nightsciencepod.bsky.social
15.05.2025 13:53 — 👍 21 🔁 5 💬 0 📌 0
Kind words from one of the up-and-coming leaders of the field ;)
14.05.2025 12:36 — 👍 0 🔁 0 💬 0 📌 0
Thanks @colmr.bsky.social ! I felt this was a suitable place to recognize the substantial efforts that go into perturbation experiments.
14.05.2025 12:34 — 👍 1 🔁 0 💬 0 📌 0
I appreciate the recognition! I guess pertomics mostly describes a reverse genetics view and "buffomics" would place the phenotype in the center ;)
14.05.2025 12:29 — 👍 0 🔁 0 💬 0 📌 0
Dynamics of Living Systems (https://www.frohlichlab.com) group leader @thecrick.bsky.social Understanding signalling & cell state dynamics through mathematical modelling and machine learning.
NCMBM, hosted at the University of Oslo, is an international research centre in interdisciplinary molecular biosciences and medicine research. NCMBM is the Norwegian node of the Nordic EMBL Partnership for Molecular Medicine.
www.med.uio.no/ncmbm/english
How do molecules and cells create life? Follow @biozentrum.unibas.ch for the latest news on research & education.
Life scientist.
Head of EMBO Membership & Elections and Courses & Workshops @embo.org #EMBOevents
previously: Editor at Molecular Systems Biology, EMBO Press
Views my own
Physician & biostatistician. Excited about cancer research, tumor evolution & functional genomics, driven to improve cancer therapy
Computational methods for epigenetic, CRISPR genome editing and single-cell genomics. Associate Professor at MGH / Harvard Medical School. http://pinellolab.org
Chief Editor of Nature Biomedical Engineering. Ph.D. Previously at Nature Methods. @rita_strack on the place formerly known as twitter.
Committed to the daily re-imagining of what a university press can be since 1962.
Website: https://mitpress.mit.edu // The Reader (our home for excerpts, essays, & interviews): https://thereader.mitpress.mit.edu
Our publications, research, and higher education solutions spread knowledge, spark curiosity and aid understanding around the world.
View our social media commenting policy here: https://cup.org/38e0Gv2
Die Alexander von Humboldt-Stiftung fördert Wissenschaftskooperationen zwischen exzellenten ausländischen und deutschen Forschenden. https://www.humboldt-foundation.de
💜-lich Willkommen auf dem offiziellen Account der Uniklinik Freiburg
Homepage: https://www.uniklinik-freiburg.de/de.html
Impressum: https://t1p.de/exco2
Division of Translational Pediatric Sarcoma Research @DKFZ
Nature Genetics is a monthly journal publishing high impact research in genetics and genomics. Part of @natureportfolio.bsky.social
Repost/like=interesting/relevant, not necessarily endorsement.
Assistant Professor, BME, Columbia University
Assistant Professor, Dep. Biology, Centre for Applied Synthetic Biology, Centre for Structural and Functional Genomics, School of Health, Concordia University | Adjunct Professor, Dep. Human Genetics, Goodman Cancer Institute, McGill University
Group leader
@sangerinstitute.bsky.social Neural diversity, spatial transcriptomics, glia, GBM
Head of Statistical Genetics, Boehringer Ingelheim
My lab at Stanford studies human population genetics and complex traits.
Assistant Professor of Molecular Genetics at the University of Toronto's Donnelly Centre
Happy European human geneticist enjoying life in the United Kingdom. Love everything genomics, even married a genomicist..;)