Are you ready for a symposium dedicated to microtubule dynamics, function, and structure? 🙌
#EESMicrotubules will bring together researchers who share an interest in microtubule biology and its relevance to health and disease 🩺
💻 s.embl.org/ees26-09-bl
✒️ Submit your abstract by 11 March
11.09.2025 14:13 — 👍 13 🔁 10 💬 0 📌 0
Microtubule end stabilisation by cooperative oligomers of Ska and Ndc80 complexes
During mitosis, properly aligned chromosomes stabilise microtubule ends with the help of kinetochores to ensure timely segregation of chromosomes. Microtubule-binding components of the human outer kinetochore, such as Ndc80 and Ska complexes, are present in multiple copies and together bind several microtubule ends, creating a highly multivalent binding interface. Whereas Ndc80:Ndc80 and Ndc80:microtubule binding is crucial for interface stability, Ndc80 alone in absence of Ska is unable to support stable kinetochore-attachments. Using cryoET, we demonstrate that oligomeric Ndc80:Ska assemblies stabilise microtubule ends against shortening by strengthening lateral contacts between tubulin protofilaments at microtubule plus-ends. We further identify a point mutation within the SKA1 microtubule-binding domain that does not affect microtubule-binding of individual Ska molecules, but does abolish Ska:Ska interactions. Finally, we report that oligomerisation of Ska, in a cooperative fashion together with the Ndc80, is necessary to maintain stable microtubule attachments both in vivo and in vitro. ### Competing Interest Statement The authors have declared no competing interest. BBSRC, BB/X014975/1, BB/W019698/1 Wellcome Trust, https://ror.org/029chgv08, 308895/Z/23/Z
New preprint from the lab - the first paper made (almost) entirely here @qmul.bsky.social
We report how human outer kinetochore complexes Ndc80 and Ska form cooperative oligomers, that together stabilise microtubule ends against shortening.
www.biorxiv.org/content/10.1...
Key results below: (1/7)
07.07.2025 09:39 — 👍 26 🔁 8 💬 1 📌 1
Excited to share our latest work in @pnas.org! Combining simulations and cryo-ET, we reveal how microtubules decide between growth and shortening. doi.org/10.1073/pnas... @maksimkalutskii.bsky.social @vladimirvolkov.bsky.social @compbiophys.bsky.social @mpi-nat.bsky.social @qmul.ac.uk @dundee.ac.uk.
03.06.2025 09:07 — 👍 34 🔁 11 💬 0 📌 3
Cryogenic light microscopy of vitrified samples with Angstrom precision https://www.biorxiv.org/content/10.1101/2025.05.27.656160v1
27.05.2025 11:48 — 👍 3 🔁 2 💬 0 📌 0
New release: napari-omero 0.5.0 😀@openmicroscopy.org @napari.org thanks to some big contributions from @psobolewskiphd.bsky.social , @will-j-moore.bsky.social and of course @talley.codes. Some features in spotlight 👇
First: napari-omero now supports lazy loading of image pyramids from OMERO🔺:
12.05.2025 11:19 — 👍 32 🔁 10 💬 1 📌 1
Enhanced axonal transport in large vertebrates: KIF5A adaptations in giraffes and pythons https://www.biorxiv.org/content/10.1101/2025.05.04.652084v1
05.05.2025 07:47 — 👍 4 🔁 2 💬 0 📌 0
This time-lapse captures 17 hours of axonal growth from a chicken dorsal root ganglion explant, visualized through the actin cytoskeleton using live confocal imaging.
I just submitted this video to the Nikon Small World in Motion competition. Today is the last day to upload yours! 😉
🧪
30.04.2025 07:16 — 👍 1422 🔁 226 💬 68 📌 26
shortest title
02.05.2025 11:58 — 👍 0 🔁 0 💬 0 📌 0
Excited to share our preprint on the molecular architecture of heterochromatin in human cells 🧬🔬w/ @jpkreysing.bsky.social, @johannesbetz.bsky.social,
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
11.04.2025 08:35 — 👍 360 🔁 142 💬 12 📌 20
Thanks to Hanjin Liu, napari now has a command palette! 🎨 This thing is really nice to use — the core team can no longer live without it — and it even works with plugin commands! 🤩
11.04.2025 10:33 — 👍 10 🔁 5 💬 1 📌 0
napari 0.6.0 — napari
It's taking us some time but it's gonna be worth the wait we promise! 😅 napari 0.6.0a1 is out now! You can install it with `pip install "napari>=0.6.0a1"`. The full release notes are here:
https://napari.org/0.6.0/release/release_0_6_0.html
But to save you a click here's a short thread: 👇
11.04.2025 10:28 — 👍 3 🔁 4 💬 1 📌 0
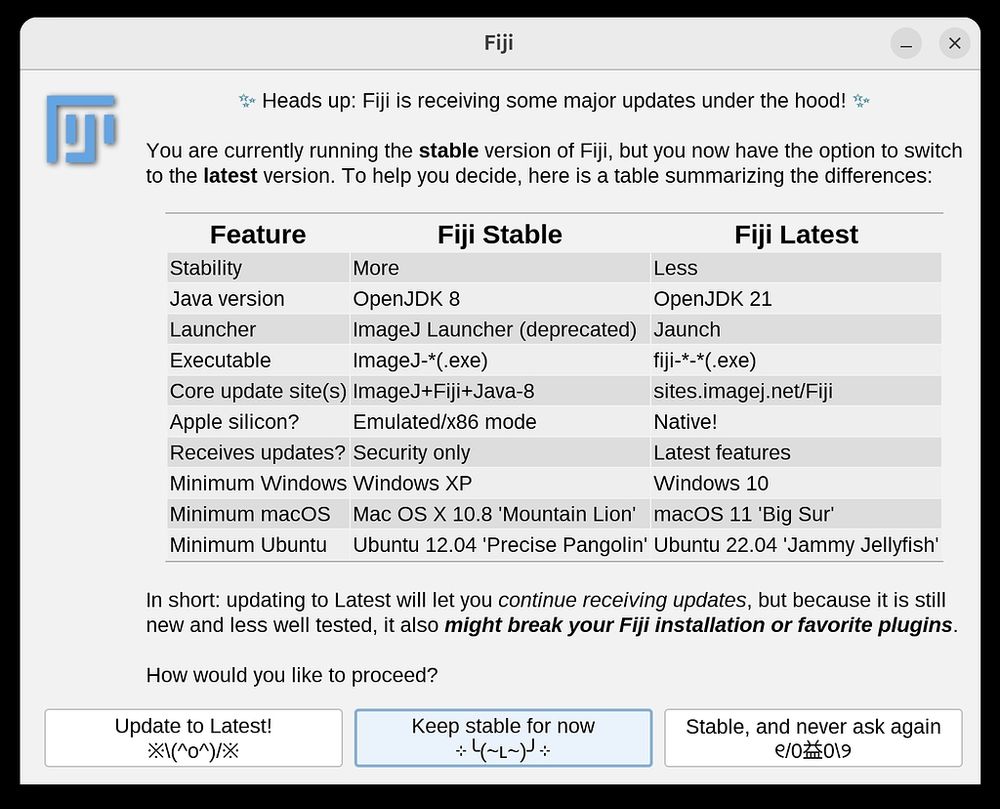
#CryoSPARC v4.7 is out! 🚀
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
09.04.2025 14:14 — 👍 52 🔁 19 💬 1 📌 2
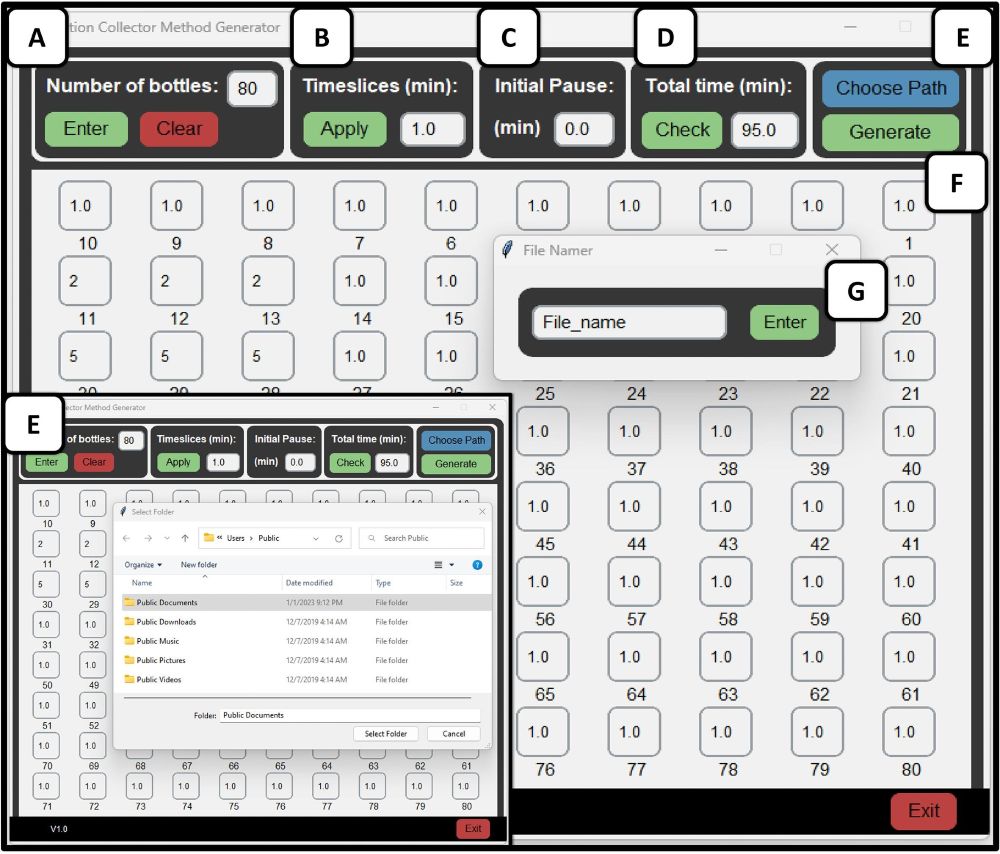
3D manual picking tool along microtubules
23.03.2025 06:27 — 👍 12 🔁 2 💬 0 📌 0

openRxiv has arrived!
We’re thrilled to announce the launch of openRxiv as an independent, researcher-led nonprofit to oversee bioRxiv and medRxiv, the world’s leading preprint servers for life and health sciences.
openrxiv.org/introducing-...
#openRxiv #OpenScience #Preprints #bioRxiv #medRxiv
11.03.2025 13:18 — 👍 406 🔁 232 💬 3 📌 19
Even though I don’t usually use it, I cannot emphasize more how much ImageJ is educational. It is a great tool to show students how image processing and image analysis works.
01.03.2025 15:48 — 👍 0 🔁 0 💬 0 📌 0
"himena" is an *infinitely extensible* application for data science I made recently.
github.com/hanjinliu/hi...
You can open any data, integrate any widget, and your plugin will look as if it is a built-in function.
Attached GIF shows my own plugin "himena-image" for image analysis 👉
13.02.2025 15:18 — 👍 0 🔁 0 💬 0 📌 0
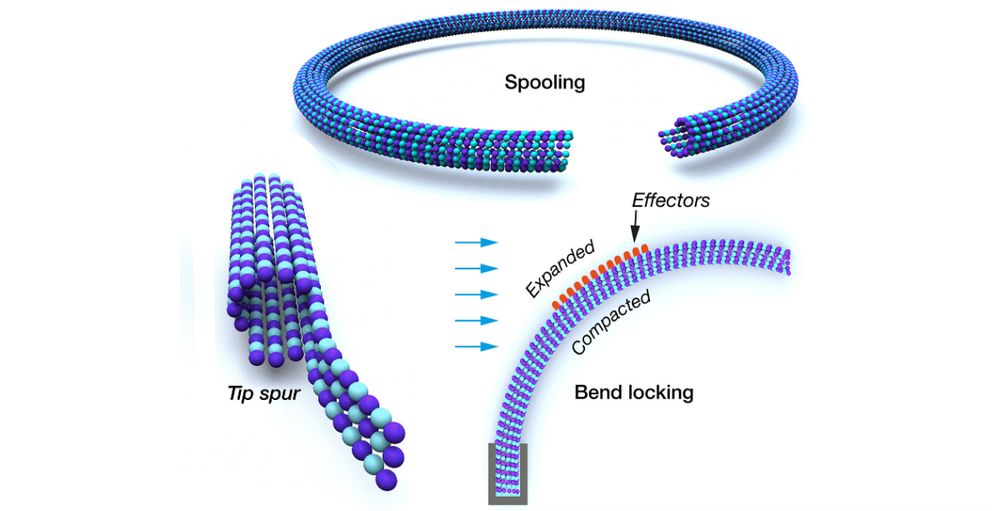
Yean Ming Chew and I wrote doi.org/10.1042/BST2... about MT lattice switching. We discuss evidence that protofilament-level structural switching is ancient and fundamental and conserved; and that tubulation creates new allosteric interfaces that operate on essentially the same structural switch.
12.02.2025 16:21 — 👍 9 🔁 5 💬 0 📌 1
Hi ChimeraX users!
CliX, the intelligent command line interface, now works very nicely with the system dark theme!
Check it out -> cxtoolshed.rbvi.ucsf.edu/apps/chimera...
06.02.2025 11:53 — 👍 1 🔁 0 💬 0 📌 0
From napari to the world: how we generalized the `conda/constructor` stack for distributing Python applications
Our work for the napari project resulted in multiple beneficial side effects for the conda packaging ecosystem.
📦 Packaging Python apps just got easier! @rjai.me 's latest #QuansightLabs blog explores how napari's team reimagined installers, creating tools that benefit the entire scientific computing community. https://buff.ly/4jMCZxx 🔬 #OpenSourceInnovation
04.02.2025 17:00 — 👍 26 🔁 8 💬 0 📌 1

We are pleased to introduce the newest members of the JCB Editorial Board: Anna Akhmanova, @msubrandizzilab.bsky.social, @goleylab.bsky.social, @christlet.bsky.social, @olzmannlab.bsky.social, Marisa Otegui, Christian Ungermann, Chonglin Yang, and Yan Zhao. https://buff.ly/3Q1O0NT
03.02.2025 17:44 — 👍 102 🔁 20 💬 1 📌 3
Fascinated by the regulatory logic behind cell phenotype-environment interactions: How biological cells leverage the structural information encoded in protein molecules to morph themselves into shapes and sizes conferring environmental stress resistance.
Post-doc in Bio-image analysis @PoL, TU Dresden, Physicist, Python code-jockey, train enthusiast.
Postdoctoral researcher at MRC Laboratory of Molecular Biology | cellular cryoET & software | Ais aiscryoet.org | Pom cryopom.streamlit.app | 😺
PostDoc @ Surrey Lab, CRG Barcelona
n-dimensional image viewer for Python
Theoretical and computational biophysics | Postdoc at Hummer's lab
@hummerlab.bsky.social | PhD at @mpibp.bsky.social | MSc. @unistuttgart.bsky.social | Physics at @uniandes.bsky.social 🇨🇴 🇩🇪 🇪🇺
Postdoc at STRUBI. Using cryo-EM to see viruses better.
Microtubule and electron optics enthusiast. PhD student at TU Delft & Leiden university. Let's make some Open Hardware.
nemoandrea.com
#Phages, #RNA, #SynBio, #DIYbio, #OpenSource, #CitizenPharma, #CompChem, #Bioinformatics, cold adaptation, #OOL, #microfluidics, #DistributedComputing, #chilis, #bass, #democracy, #PIRATEN
Fediverse: @MWNautilus@mstdn.social
Studying cytoskeleton and mitosis, focused on tubulin and microtubules. Curious about bacterial division and Ftsz. Interested in computational modeling, all kinds of microscopy, experimental and theoretical biophysics, bio- and med- chemisty
Virginie Hamel & Paul Guichard Lab at University of Geneva
#cryoEM/ET❄️ and #UExM ⚗️ #ExpansionMicroscopy #TeamTomo
Genève, Suisse 🇨🇭
https://mocel.unige.ch/research-groups/guichard-hamel/overv
Postdoc at Maeshima-Lab (@kazu-maeshima.bsky.social), National Institute of Genetics, Japan / Interested in chromatin, replication, microscopy, ...
Postdoc @Moores Lab in London. Interested in cytoskeleton and cryoEM🐍🐍🐍
napari community manager and core dev, developmental neuroscientist, birder, and the proudest papa.
I am a Software Architect for AI Solutions, Python Fan, Microscopist Image Analyst and really like Skiing.
Science is like magic but real.
Postdoctoral Researcher at BDR, RIKEN
Nonequilibrium physics of living matter Riken Hakubi research team.
Twitter: https://twitter.com/fukaity
HP: https://yfukai.net/
Microscopist and Bio-Image Analyst for the BioImaging and Optics Platform https://biop.epfl.ch/ at EPFL
https://nicolas.chiaruttini.org
Professor at UTokyo. (postdoc @Stanford) / b.1986 / structural biology, protein engineering, rhodopsin, GPCRs http://tinyurl.com/ycyf87t7