Congrats Jenny! 🥳
19.08.2025 07:01 — 👍 1 🔁 0 💬 1 📌 0
Thank you 😊
13.08.2025 09:06 — 👍 0 🔁 0 💬 0 📌 0
Thank you! It's EMC time 😉
12.08.2025 07:02 — 👍 1 🔁 0 💬 0 📌 0
Thank you to everyone involved in the Schulman @mpibiochem.bsky.social , Feige @tum.de , @fenech-lab.bsky.social , and Schuldiner labs! 🥳
11.08.2025 12:35 — 👍 0 🔁 0 💬 0 📌 0

A potential model: the conserved EMC:Spf1 supercomplex spatially couples insertion and extraction. Juxtaposed functional sites form a shared cavity, enabling substrate handover and discrimination. Spf1's nucleotide state may regulate access to this cavity, coordinating insertion and extraction.
11.08.2025 12:35 — 👍 0 🔁 0 💬 1 📌 0

Translocation by Spf1 is coupled to ATP hydrolysis. To probe its functional cycle in the EMC:Spf1 complex, we determined structures in the E1-ATP and E1-P states.
ATP binding stabilizes Spf1’s “arm” domain, contacting EMC’s cytoplasmic cap above the insertase cavity, closing the composite cavity.
11.08.2025 12:35 — 👍 1 🔁 0 💬 1 📌 0

The high stability of the yeast EMC:Spf1 complex suggests a key functional relationship. Using endogenous tagging, mass spectrometry, modeling and experimental validation, we show that a similar complex exists in human cells between EMC and ATP13A1.
11.08.2025 12:35 — 👍 0 🔁 0 💬 1 📌 0

The main site of interaction is confined to a lumenal interface (“lumenal dock”) involving EMC7, EMC10, and the charged lumenal surface of Spf1.
11.08.2025 12:35 — 👍 0 🔁 0 💬 1 📌 0

The architecture of this supercomplex reveals juxtaposed functional sites for TMD insertion (EMC) and extraction (Spf1), forming a large composite intramembrane cavity.
11.08.2025 12:35 — 👍 2 🔁 0 💬 1 📌 0

We found that at endogenous levels in yeast, the EMC forms a stoichiometric complex with Spf1. Spf1 is a TMD dislocase, the biochemical counterpart to EMC's role as insertase. To gain more insights into this intriguing supercomplex, we determined the EMC:Spf1 structure by cryo-EM.
11.08.2025 12:35 — 👍 0 🔁 0 💬 1 📌 0
Huge thanks to all co-authors at @tum.de and @mpibiochem.bsky.social for making this work possible!
#ERliterature #chaperone #proteostasis
9/9
05.08.2025 13:18 — 👍 1 🔁 0 💬 0 📌 0
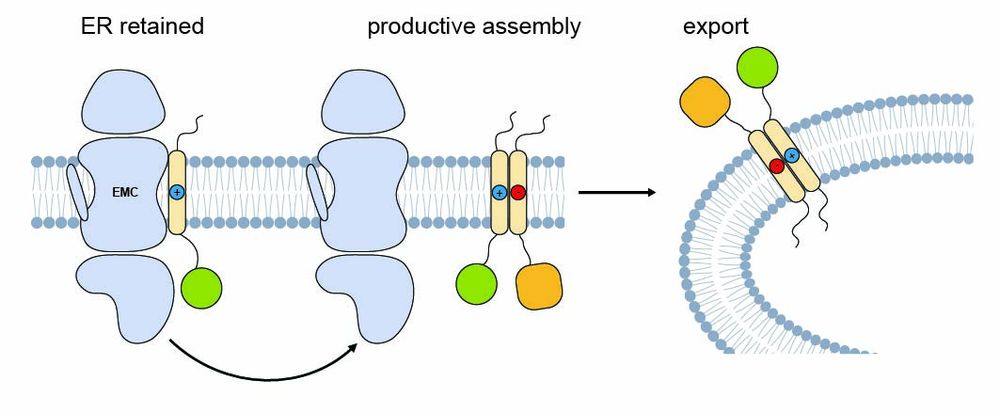
So what happens after binding?
We found that challenging TMDs remain bound to EMC and are ER-retained—but once a partner for productive assembly is available, EMC binding is reduced and the protein can exit the ER.
8/9
05.08.2025 13:18 — 👍 1 🔁 0 💬 1 📌 0
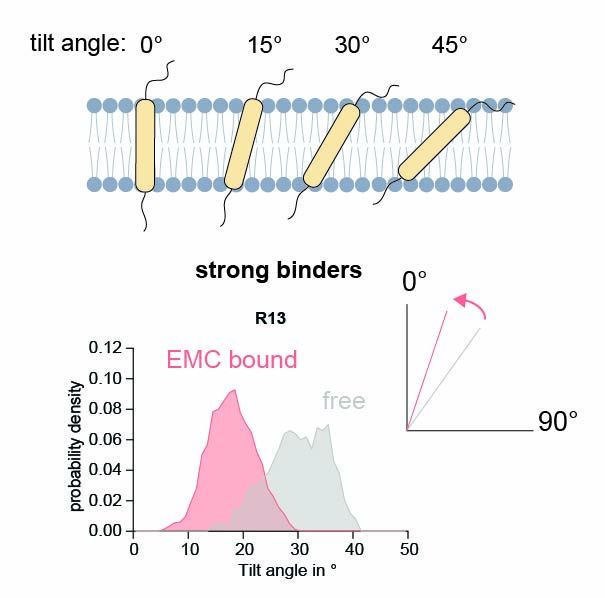
Molecular dynamics simulations explain this: Polar residues induce a tilted orientation of the TMD in the bilayer. EMC binding stabilizes them in an upright pose, likely facilitating proper folding and assembly.
7/9
05.08.2025 13:18 — 👍 0 🔁 0 💬 1 📌 0
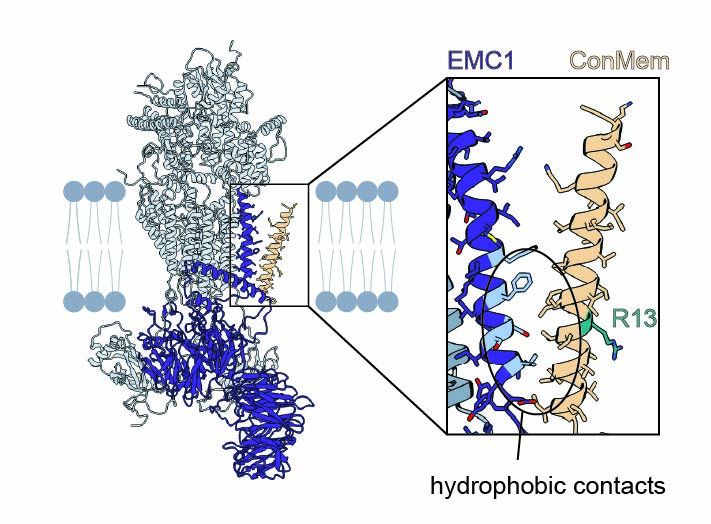
But how does EMC recognize them?
Surprisingly, mutational analysis and site-specific crosslinking showed that EMC doesn't bind the polar face of the TMD—but engages the opposite, hydrophobic side.
6/9
05.08.2025 13:18 — 👍 0 🔁 0 💬 1 📌 0
Why these clients?
Their TMDs often contain polar/charged residues needed for function but are only marginally stable in the membrane—making them ideal candidates for chaperone support during folding and assembly.
5/9
05.08.2025 13:18 — 👍 0 🔁 0 💬 1 📌 0
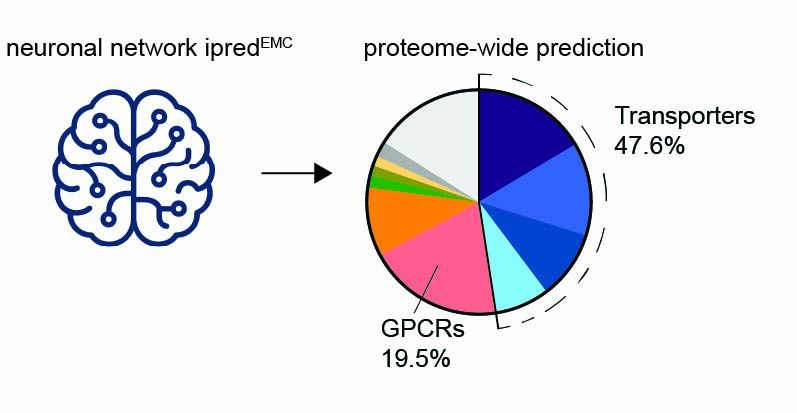
How does this translate to natural proteins?
We trained and validated a neural network (ipredEMC) to predict EMC binding proteome-wide. This tool revealed that transporters and ion channels are major chaperone clients.
4/9
05.08.2025 13:18 — 👍 0 🔁 0 💬 1 📌 0
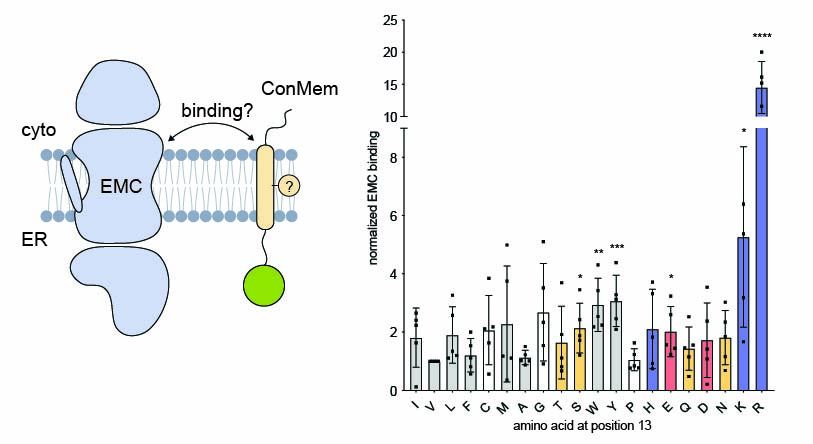
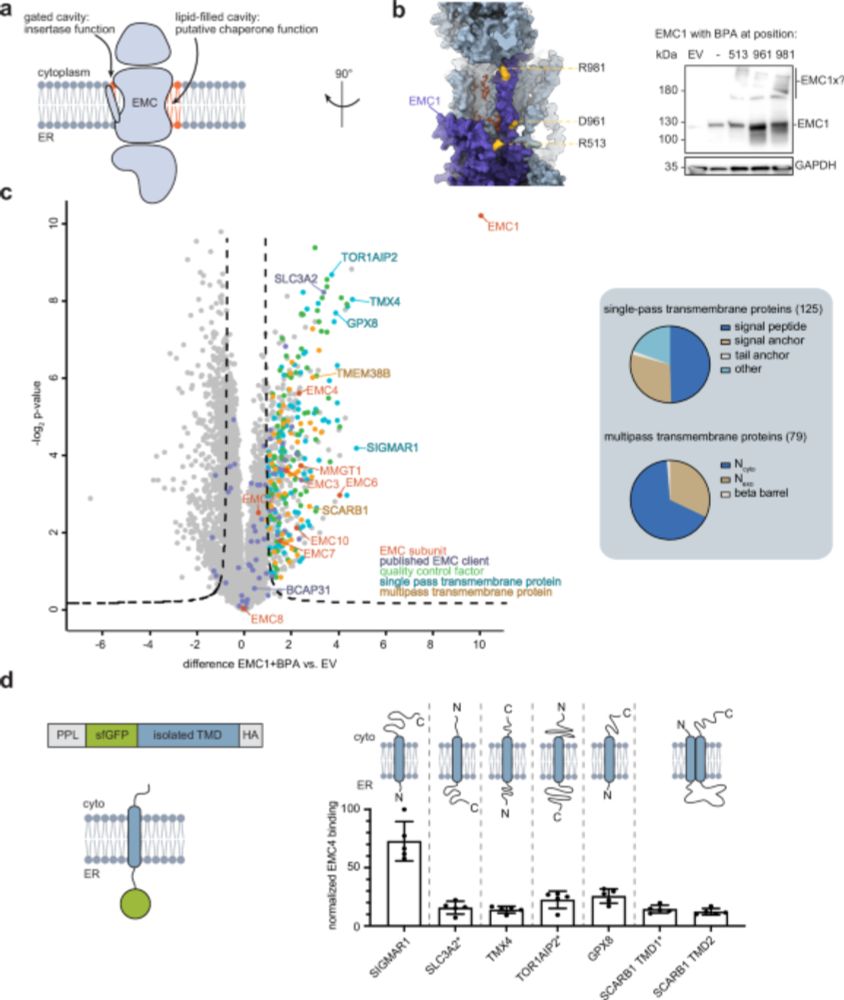
To understand what drives EMC binding, we turned to a minimal model system. Using a single-pass model transmembrane domain (TMD) and systematic residue substitutions, we found that mostly polar and charged residues within the TMD enhace EMC binding.
3/9
05.08.2025 13:18 — 👍 0 🔁 0 💬 1 📌 0
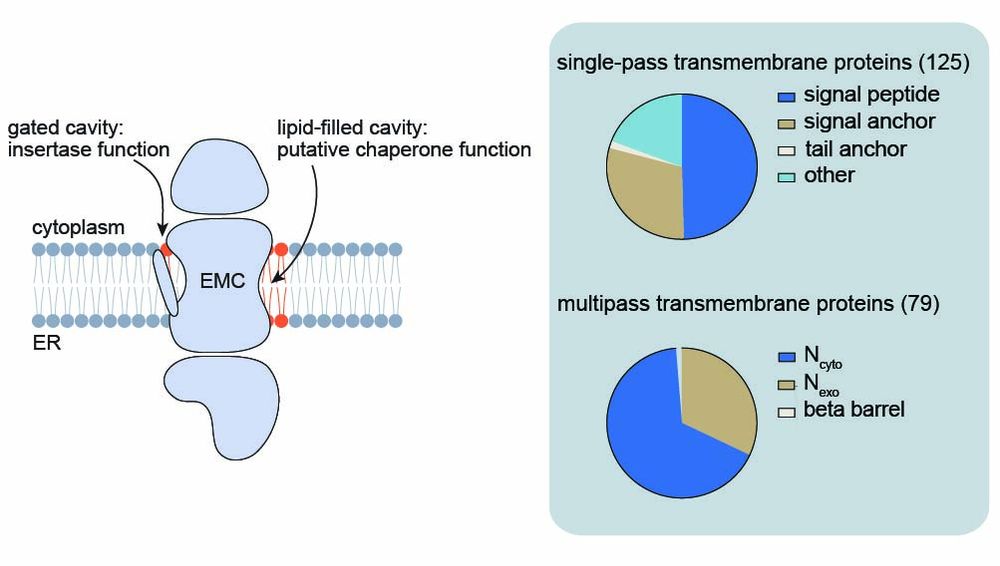
Using site-specific photocrosslinking and mass spectrometry, we mapped interactions at the lipid-filled cavity of the EMC, revealing a broad spectrum of membrane proteins extending far beyond known insertase clients.
2/9
05.08.2025 13:18 — 👍 0 🔁 0 💬 1 📌 0
Congratulations Leo! 🥳
24.03.2025 15:25 — 👍 0 🔁 0 💬 1 📌 0
Wow, that's amazing! Congratulations!
05.03.2025 18:42 — 👍 1 🔁 0 💬 1 📌 0
Structural biology on DNA repair, chromatin remodelling and innate immunity. Located at the Gene Center of the LMU in Munich.
https://www.genzentrum.uni-muenchen.de/research-groups/hopfner/index.html
enthusiast of transposable elements, genetic conflicts, small RNAs, Drosophila, and funky germline biology
running a lab at IMBA, Vienna BioCenter
https://www.oeaw.ac.at/imba/research/julius-brennecke
Cell biologist at UNC interested in organelles, lipids, neurodegenerative disease, and light microscopy. https://cohenlaboratory.web.unc.edu
PhD student at the Brennecke and Plaschka labs, Vienna.
Interested in RNA silencing
IMPRS-ML PhD Candidate in the Briggs and Schulman lab @briggsgroup.bsky.social @mpibiochem.bsky.social | #cryo-ET #CLEM
Molecular biologist interested in proteostasis and neurodegeneration | Postdoc at Max Planck Institute Biochemistry
Shooting electrons, ions and photons (mainly) at plants to study cell-cell communication @mpibiochem.bsky.social & @hhu.de
Postdoc with Sascha Martens and Tim Clausen at the Vienna BioCenter (VBC). #MSCA Fellow. Formerly University of Oxford and University of Antwerp.
CryoEM, membrane proteins and whatnot
❄️🔬 cryoEM/ET and subtomogram averaging | Mitochondria are the powerhouse of the cell ⚡️ | PhD Student @uniGoettingen.bsky.social #teamtomo
IMPRS-ML student in Schulman lab, Max Planck Institute of Biochemistry🧪 BIF PhD Fellow 🔬 Interested in Ubiquitin, Protein Degradation and Molecular Signaling 👩🏻🔬
Protein quality control and stress response lab at the Center for Molecular Medicine Cologne (CMMC) and CECAD research center
https://www.trentini-lab.cmmc-uni-koeln.de/
(Icon made by Freepik from www.flaticon.com)
Scientist. She/her. I like cryoET, fluorescences microscopy, and cell biology.
Developing cryoCLEM methods in the Mahamid lab at EMBL Heidelberg. Formerly studying NPCs in the Beck Lab at MPIBP Frankfurt. MBL Physiology’23 alumn.
IMPRS-ML PhD Student in the Schulman Lab at Max Planck Institute of Biochemistry
Postdoc @SophieMartinLab.bsky.social studying #GPCR and #MAPK signalling in fission yeast. Alumnus of @mjafreeman.bsky.social and @jordanraff.bsky.social
Assistant Professor @Stanford_Molecular & Cellular Physiology; Membrane protein homeostasis; Protein quality control; Antibody engineering; Engineering cells with genetically encodable #nanobodies, #FirstGen; Pronouns: He/Him
https://www.pleinerlab.org/
Nature Communications is an open access journal publishing high-quality research in all areas of the biological, physical, chemical, clinical, social, and Earth sciences.
www.nature.com/ncomms/
Photosynthesis and cryoEM at Biozentrum, Basel.
PhD student in Schulman lab @mpibiochem.bsky.social
Navigating through biochemistry and structural biology of ubiquitin world 👩🏼🔬 🔬